Trim41 is required to regulate chromosome axis protein dynamics and meiosis in male mice
- PMID: 35648791
- PMCID: PMC9191731
- DOI: 10.1371/journal.pgen.1010241
Trim41 is required to regulate chromosome axis protein dynamics and meiosis in male mice
Abstract
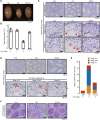
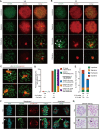
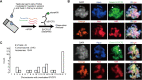
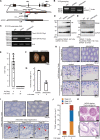
Meiosis is a hallmark event in germ cell development that accompanies sequential events executed by numerous molecules. Therefore, characterization of these factors is one of the best strategies to clarify the mechanism of meiosis. Here, we report tripartite motif-containing 41 (TRIM41), a ubiquitin ligase E3, as an essential factor for proper meiotic progression and fertility in male mice. Trim41 knockout (KO) spermatocytes exhibited synaptonemal complex protein 3 (SYCP3) overloading, especially on the X chromosome. Furthermore, mutant mice lacking the RING domain of TRIM41, required for the ubiquitin ligase E3 activity, phenocopied Trim41 KO mice. We then examined the behavior of mutant TRIM41 (ΔRING-TRIM41) and found that ΔRING-TRIM41 accumulated on the chromosome axes with overloaded SYCP3. This result suggested that TRIM41 exerts its function on the chromosome axes. Our study revealed that Trim41 is essential for preventing SYCP3 overloading, suggesting a TRIM41-mediated mechanism for regulating chromosome axis protein dynamics during male meiotic progression.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures


Similar articles
-
Deciphering meiotic chromatin organization by SYCP3.Nucleic Acids Res. 2025 Jun 6;53(11):gkaf460. doi: 10.1093/nar/gkaf460. Nucleic Acids Res. 2025. PMID: 40488283 Free PMC article.
-
Mouse SYCP2 is required for synaptonemal complex assembly and chromosomal synapsis during male meiosis.J Cell Biol. 2006 May 22;173(4):497-507. doi: 10.1083/jcb.200603063. J Cell Biol. 2006. PMID: 16717126 Free PMC article.
-
Meiotic cohesin STAG3 is required for chromosome axis formation and sister chromatid cohesion.EMBO J. 2014 Jun 2;33(11):1256-70. doi: 10.1002/embj.201387330. Epub 2014 May 5. EMBO J. 2014. PMID: 24797474 Free PMC article.
-
Live cell analyses of synaptonemal complex dynamics and chromosome movements in cultured mouse testis tubules and embryonic ovaries.Chromosoma. 2018 Sep;127(3):341-359. doi: 10.1007/s00412-018-0668-7. Epub 2018 Mar 26. Chromosoma. 2018. PMID: 29582139 Free PMC article.
-
The role of spermatogonially expressed germ cell-specific genes in mammalian meiosis.Chromosome Res. 2007;15(5):623-32. doi: 10.1007/s10577-007-1141-2. Chromosome Res. 2007. PMID: 17674150 Review.
Cited by
-
Heat shock factor 5 establishes the male germ-line meiotic sex chromosome inactivation through regulation of Smarca4.Heliyon. 2023 Apr 21;9(5):e15194. doi: 10.1016/j.heliyon.2023.e15194. eCollection 2023 May. Heliyon. 2023. Retraction in: Heliyon. 2025 Apr 02;11(9):e43282. doi: 10.1016/j.heliyon.2025.e43282. PMID: 37206036 Free PMC article. Retracted.
References
-
- Versteeg GA, Rajsbaum R, Sánchez-Aparicio MT, Maestre AM, Valdiviezo J, Shi M, et al.. The E3-ligase TRIM family of proteins regulates signaling pathways triggered by innate immune pattern-recognition receptors. Immunity. 2013;38(2):384–98. Epub 2013/02/27. doi: 10.1016/j.immuni.2012.11.013 ; PubMed Central PMCID: PMC3584420. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

