Synergistic autoinhibition and activation mechanisms control kinesin-1 motor activity
- PMID: 35649356
- PMCID: PMC9365671
- DOI: 10.1016/j.celrep.2022.110900
Synergistic autoinhibition and activation mechanisms control kinesin-1 motor activity
Erratum in
-
Synergistic autoinhibition and activation mechanisms control kinesin-1 motor activity.Cell Rep. 2022 Jun 28;39(13):111016. doi: 10.1016/j.celrep.2022.111016. Cell Rep. 2022. PMID: 35767955 Free PMC article. No abstract available.
Abstract
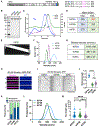
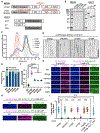
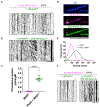
Kinesin-1 activity is regulated by autoinhibition. Intramolecular interactions within the kinesin heavy chain (KHC) are proposed to be one facet of motor regulation. The KHC also binds to the kinesin light chain (KLC), which has been implicated in both autoinhibition and activation of the motor. We show that the KLC inhibits the kinesin-microtubule interaction independently from the proposed intramolecular interaction within KHC. Cargo-adaptor proteins that bind the KLC stimulated processive movement, but the landing rate of activated kinesin complexes remained low. Mitogen-activated protein 7 (MAP7) enhanced motility by increasing the landing rate and run length of the activated kinesin motors. Our results support a model whereby the motor activity of the kinesin is regulated by synergistic inhibition mechanisms and that cargo-adaptor binding to the KLC releases both mechanisms. However, a non-motor MAP is required for robust microtubule association of the activated motor. Thus, human kinesin is regulated by synergistic autoinhibition and activation mechanisms.
Keywords: CP: Molecular biology; dynein; kinesin; microtubule; motor protein.
Copyright © 2022 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures


References
-
- Crimella C, Baschirotto C, Arnoldi A, Tonelli A, Tenderini E, Airoldi G, Martinuzzi A, Trabacca A, Losito L, Scarlato M, et al. (2012). Mutations in the motor and stalk domains of KIF5A in spastic paraplegia type 10 and in axonal Charcot-Marie-Tooth type 2. Clin. Genet 82, 157–164. 10.1111/j.1399-0004.2011.01717.x. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

