KLFDAPC: a supervised machine learning approach for spatial genetic structure analysis
- PMID: 35649387
- PMCID: PMC9294434
- DOI: 10.1093/bib/bbac202
KLFDAPC: a supervised machine learning approach for spatial genetic structure analysis
Abstract
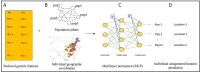
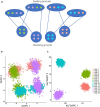
Geographic patterns of human genetic variation provide important insights into human evolution and disease. A commonly used tool to detect and describe them is principal component analysis (PCA) or the supervised linear discriminant analysis of principal components (DAPC). However, genetic features produced from both approaches could fail to correctly characterize population structure for complex scenarios involving admixture. In this study, we introduce Kernel Local Fisher Discriminant Analysis of Principal Components (KLFDAPC), a supervised non-linear approach for inferring individual geographic genetic structure that could rectify the limitations of these approaches by preserving the multimodal space of samples. We tested the power of KLFDAPC to infer population structure and to predict individual geographic origin using neural networks. Simulation results showed that KLFDAPC has higher discriminatory power than PCA and DAPC. The application of our method to empirical European and East Asian genome-wide genetic datasets indicated that the first two reduced features of KLFDAPC correctly recapitulated the geography of individuals and significantly improved the accuracy of predicting individual geographic origin when compared to PCA and DAPC. Therefore, KLFDAPC can be useful for geographic ancestry inference, design of genome scans and correction for spatial stratification in GWAS that link genes to adaptation or disease susceptibility.
Keywords: individual geographic origin; machine learning; population structure.
© The Author(s) 2022. Published by Oxford University Press.
Figures




References
-
- Barbujani G, Excoffier LGL. The history and geography of human genetic diversity. In: Stearns, Stephen C. (Ed.). Evolution in health and disease. Oxford: Oxford University Press, 1999. https://archive-ouverte.unige.ch/unige:93149.
-
- Labonte R, Polanyi M, Muhajarine N, et al. Beyond the divides: towards critical population health research. Crit Public Health 2005;15:5–17.
-
- Parsons T. Societies: Evolutionary and Comparative Perspectives. Englewood Cliffs, NJ: Prentice-Hall, 1966.
-
- Root M. How we divide the world. Philos Sci 2000;67:S628–39.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

