The androgen receptor is a therapeutic target in desmoplastic small round cell sarcoma
- PMID: 35650195
- PMCID: PMC9160255
- DOI: 10.1038/s41467-022-30710-z
The androgen receptor is a therapeutic target in desmoplastic small round cell sarcoma
Abstract
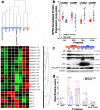
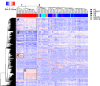
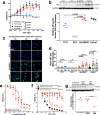
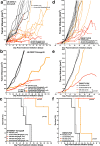
Desmoplastic small round cell tumor (DSRCT) is an aggressive, usually incurable sarcoma subtype that predominantly occurs in post-pubertal young males. Recent evidence suggests that the androgen receptor (AR) can promote tumor progression in DSRCTs. However, the mechanism of AR-induced oncogenic stimulation remains undetermined. Herein, we demonstrate that enzalutamide and AR-directed antisense oligonucleotides (AR-ASO) block 5α-dihydrotestosterone (DHT)-induced DSRCT cell proliferation and reduce xenograft tumor burden. Gene expression analysis and chromatin immunoprecipitation sequencing (ChIP-seq) were performed to elucidate how AR signaling regulates cellular epigenetic programs. Remarkably, ChIP-seq revealed novel DSRCT-specific AR DNA binding sites adjacent to key oncogenic regulators, including WT1 (the C-terminal partner of the pathognomonic fusion protein) and FOXF1. Additionally, AR occupied enhancer sites that regulate the Wnt pathway, neural differentiation, and embryonic organ development, implicating AR in dysfunctional cell lineage commitment. Our findings have direct clinical implications given the widespread availability of FDA-approved androgen-targeted agents used for prostate cancer.
© 2022. The Author(s).
Conflict of interest statement
A.R.M. is an employee and shareholder of Ionis Pharmaceuticals. The remaining authors declare no competing interests.
Figures



References
-
- Honore, C. et al. Abdominal desmoplastic small round cell tumor: multimodal treatment combining chemotherapy, surgery, and radiotherapy is the best option. Ann. Surg. Oncol.22, 1073–1079. 10.1245/s10434-014-4123-6 (2015). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

