CoV2K model, a comprehensive representation of SARS-CoV-2 knowledge and data interplay
- PMID: 35650205
- PMCID: PMC9160032
- DOI: 10.1038/s41597-022-01348-9
CoV2K model, a comprehensive representation of SARS-CoV-2 knowledge and data interplay
Abstract
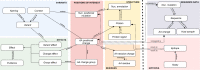
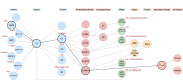
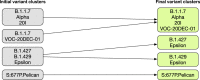
Since the outbreak of the COVID-19 pandemic, many research organizations have studied the genome of the SARS-CoV-2 virus; a body of public resources have been published for monitoring its evolution. While we experience an unprecedented richness of information in this domain, we also ascertained the presence of several information quality issues. We hereby propose CoV2K, an abstract model for explaining SARS-CoV-2-related concepts and interactions, focusing on viral mutations, their co-occurrence within variants, and their effects. CoV2K provides a clear and concise route map for understanding different connected types of information related to the virus; it thus drives a process of data and knowledge integration that aggregates information from several current resources, harmonizing their content and overcoming incompleteness and inconsistency issues. CoV2K is available for exploration as a graph that can be queried through a RESTful API addressing single entities or paths through their relationships. Practical use cases demonstrate its application to current knowledge inquiries.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- World Health Organization. Tracking SARS-CoV-2 variants. https://www.who.int/en/activities/tracking-SARS-CoV-2-variants/. Last accessed: March 8th, 2022. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

