Nuclear Vav3 is required for polycomb repression complex-1 activity in B-cell lymphoblastic leukemogenesis
- PMID: 35650206
- PMCID: PMC9160250
- DOI: 10.1038/s41467-022-30651-7
Nuclear Vav3 is required for polycomb repression complex-1 activity in B-cell lymphoblastic leukemogenesis
Abstract
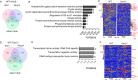
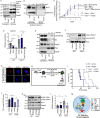
Acute B-cell lymphoblastic leukemia (B-ALL) results from oligo-clonal evolution of B-cell progenitors endowed with initiating and propagating leukemia properties. The activation of both the Rac guanine nucleotide exchange factor (Rac GEF) Vav3 and Rac GTPases is required for leukemogenesis mediated by the oncogenic fusion protein BCR-ABL. Vav3 expression becomes predominantly nuclear upon expression of BCR-ABL signature. In the nucleus, Vav3 interacts with BCR-ABL, Rac, and the polycomb repression complex (PRC) proteins Bmi1, Ring1b and Ezh2. The GEF activity of Vav3 is required for the proliferation, Bmi1-dependent B-cell progenitor self-renewal, nuclear Rac activation, protein interaction with Bmi1, mono-ubiquitination of H2A(K119) (H2AK119Ub) and repression of PRC-1 (PRC1) downstream target loci, of leukemic B-cell progenitors. Vav3 deficiency results in de-repression of negative regulators of cell proliferation and repression of oncogenic transcriptional factors. Mechanistically, we show that Vav3 prevents the Phlpp2-sensitive and Akt (S473)-dependent phosphorylation of Bmi1 on the regulatory residue S314 that, in turn, promotes the transcriptional factor reprogramming of leukemic B-cell progenitors. These results highlight the importance of non-canonical nuclear Rho GTPase signaling in leukemogenesis.
© 2022. The Author(s).
Conflict of interest statement
A.B. is a co-founder of Datirium, LLC. Datirium developed SciDAP bioinformatics platform used for data analysis in this study. Other authors have no relevant conflicts of interest to declare.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

