Leaf transcriptome profiling of contrasting sugarcane genotypes for drought tolerance under field conditions
- PMID: 35650424
- PMCID: PMC9160059
- DOI: 10.1038/s41598-022-13158-5
Leaf transcriptome profiling of contrasting sugarcane genotypes for drought tolerance under field conditions
Abstract
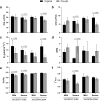
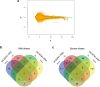
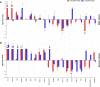
Drought is the most detrimental abiotic stress to sugarcane production. Nevertheless, transcriptomic analyses remain scarce for field-grown plants. Here we performed comparative transcriptional profiling of two contrasting sugarcane genotypes, 'IACSP97-7065' (drought-sensitive) and 'IACSP94-2094' (drought-tolerant) grown in a drought-prone environment. Physiological parameters and expression profiles were analyzed at 42 (May) and 117 (August) days after the last rainfall. The first sampling was done under mild drought (soil water potential of -60 kPa), while the second one was under severe drought (soil water potential of -75 kPa). Microarray analysis revealed a total of 622 differentially expressed genes in both sugarcane genotypes under mild and severe drought stress, uncovering about 250 exclusive transcripts to 'IACSP94-2094' involved in oxidoreductase activity, transcriptional regulation, metabolism of amino acids, and translation. Interestingly, the enhanced antioxidant system of 'IACSP94-2094' may protect photosystem II from oxidative damage, which partially ensures stable photochemical activity even after 117 days of water shortage. Moreover, the tolerant genotype shows a more extensive set of responsive transcription factors, promoting the fine-tuning of drought-related molecular pathways. These results help elucidate the intrinsic molecular mechanisms of a drought-tolerant sugarcane genotype to cope with ever-changing environments, including prolonged water deficit, and may be useful for plant breeding programs.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
Drought-triggered leaf transcriptional responses disclose key molecular pathways underlying leaf water use efficiency in sugarcane (Saccharum spp.).Front Plant Sci. 2023 May 8;14:1182461. doi: 10.3389/fpls.2023.1182461. eCollection 2023. Front Plant Sci. 2023. PMID: 37223790 Free PMC article.
-
Short-term physiological changes in roots and leaves of sugarcane varieties exposed to H2O2 in root medium.J Plant Physiol. 2015 Apr 1;177:93-99. doi: 10.1016/j.jplph.2015.01.009. Epub 2015 Jan 25. J Plant Physiol. 2015. PMID: 25703773
-
Morpho-physio-biochemical and gene expression profiling of drought-tolerant sugarcane genotypes (Saccharum hybrids).Mol Biol Rep. 2025 May 25;52(1):504. doi: 10.1007/s11033-025-10592-2. Mol Biol Rep. 2025. PMID: 40413679
-
MicroRNAs and drought responses in sugarcane.Front Plant Sci. 2015 Feb 23;6:58. doi: 10.3389/fpls.2015.00058. eCollection 2015. Front Plant Sci. 2015. PMID: 25755657 Free PMC article. Review.
-
Drought and salinity stresses induced physio-biochemical changes in sugarcane: an overview of tolerance mechanism and mitigating approaches.Front Plant Sci. 2023 Aug 11;14:1225234. doi: 10.3389/fpls.2023.1225234. eCollection 2023. Front Plant Sci. 2023. PMID: 37645467 Free PMC article. Review.
Cited by
-
Integrated PacBio SMRT and Illumina sequencing uncovers transcriptional and physiological responses to drought stress in whole-plant Nitraria tangutorum.Front Genet. 2024 Oct 1;15:1474259. doi: 10.3389/fgene.2024.1474259. eCollection 2024. Front Genet. 2024. PMID: 39411372 Free PMC article.
-
Transcriptome profiling disclosed the effect of single and combined drought and heat stress on reprogramming of genes expression in barley flag leaf.Front Plant Sci. 2023 Jan 16;13:1096685. doi: 10.3389/fpls.2022.1096685. eCollection 2022. Front Plant Sci. 2023. PMID: 36726667 Free PMC article.
-
Comparative transcriptomic analyses of two sugarcane Saccharum L. cultivars differing in drought tolerance.Front Plant Sci. 2023 Oct 11;14:1243664. doi: 10.3389/fpls.2023.1243664. eCollection 2023. Front Plant Sci. 2023. PMID: 37885666 Free PMC article.
-
Drought-triggered leaf transcriptional responses disclose key molecular pathways underlying leaf water use efficiency in sugarcane (Saccharum spp.).Front Plant Sci. 2023 May 8;14:1182461. doi: 10.3389/fpls.2023.1182461. eCollection 2023. Front Plant Sci. 2023. PMID: 37223790 Free PMC article.
References
-
- FAO. Statistical Database.https://www.fao.org/faostat/ (2021).
-
- UNICA. Sugarcane Database. https://www.unica.com.br (2021).
-
- de Andrade LM, et al. Characterization of PIP2 aquaporins in Saccharum hybrids. Plant Gene. 2016;5:31–37. doi: 10.1016/j.plgene.2015.11.004. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

