Differential cofactor dependencies define distinct types of human enhancers
- PMID: 35650434
- PMCID: PMC7613064
- DOI: 10.1038/s41586-022-04779-x
Differential cofactor dependencies define distinct types of human enhancers
Abstract
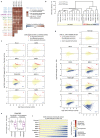
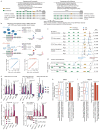
All multicellular organisms rely on differential gene transcription regulated by genomic enhancers, which function through cofactors that are recruited by transcription factors1,2. Emerging evidence suggests that not all cofactors are required at all enhancers3-5, yet whether these observations reflect more general principles or distinct types of enhancers remained unknown. Here we categorized human enhancers by their cofactor dependencies and show that these categories provide a framework to understand the sequence and chromatin diversity of enhancers and their roles in different gene-regulatory programmes. We quantified enhancer activities along the entire human genome using STARR-seq6 in HCT116 cells, following the rapid degradation of eight cofactors. This analysis identified different types of enhancers with distinct cofactor requirements, sequences and chromatin properties. Some enhancers were insensitive to the depletion of the core Mediator subunit MED14 or the bromodomain protein BRD4 and regulated distinct transcriptional programmes. In particular, canonical Mediator7 seemed dispensable for P53-responsive enhancers, and MED14-depleted cells induced endogenous P53 target genes. Similarly, BRD4 was not required for the transcription of genes that bear CCAAT boxes and a TATA box (including histone genes and LTR12 retrotransposons) or for the induction of heat-shock genes. This categorization of enhancers through cofactor dependencies reveals distinct enhancer types that can bypass broadly utilized cofactors, which illustrates how alternative ways to activate transcription separate gene expression programmes and provide a conceptual framework to understand enhancer function and regulatory specificity.
© 2022. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
The authors declare no competing interests.
Figures












Comment in
-
Cofactors: a new layer of specificity to enhancer regulation.Trends Biochem Sci. 2022 Dec;47(12):993-995. doi: 10.1016/j.tibs.2022.07.008. Epub 2022 Aug 13. Trends Biochem Sci. 2022. PMID: 35970663
-
Enhancers have more cofactor specificity than we think: A potential new way to classify enhancers based on their functional cofactor requirements.Mol Cell. 2022 Aug 18;82(16):2922-2924. doi: 10.1016/j.molcel.2022.07.015. Mol Cell. 2022. PMID: 35985301
References
-
- Reiter F, Wienerroither S, Stark A. Combinatorial function of transcription factors and cofactors. Current Opinion in Genetics & Development. 2017;43:73–81. - PubMed
-
- Nakagawa T, Yoneda M, Higashi M, Ohkuma Y, Ito T. Enhancer function regulated by combinations of transcription factors and cofactors. Genes Cells. 2018;23:808–821. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

