Degradome sequencing reveals an integrative miRNA-mediated gene interaction network regulating rice seed vigor
- PMID: 35650544
- PMCID: PMC9158300
- DOI: 10.1186/s12870-022-03645-2
Degradome sequencing reveals an integrative miRNA-mediated gene interaction network regulating rice seed vigor
Erratum in
-
Correction: Degradome sequencing reveals an integrative miRNA-mediated gene interaction network regulating rice seed vigor.BMC Plant Biol. 2022 Jun 28;22(1):312. doi: 10.1186/s12870-022-03700-y. BMC Plant Biol. 2022. PMID: 35764938 Free PMC article. No abstract available.
Abstract
Background: It is well known that seed vigor is essential for agricultural production and rice (Oryza sativa L.) is one of the most important crops in the world. Though we previously reported that miR164c regulates rice seed vigor, but whether and how other miRNAs cooperate with miR164c to regulate seed vigor is still unknown.
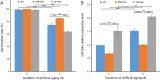
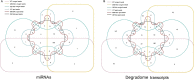
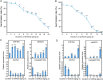
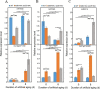
Results: Based on degradome data of six RNA samples isolated from seeds of the wild-type (WT) indica rice cultivar 'Kasalath' as well as two modified lines in 'Kasalath' background (miR164c-silenced line [MIM164c] and miR164c overexpression line [OE164c]), which were subjected to either no aging treatment or an 8-day artificial aging treatment, 1247 different target transcripts potentially cleaved by 421 miRNAs were identified. The miRNA target genes were functionally annotated via GO and KEGG enrichment analyses. By STRING database assay, a miRNA-mediated gene interaction network regulating seed vigor in rice was revealed, which comprised at least four interconnected pathways: the miR5075-mediated oxidoreductase related pathway, the plant hormone related pathway, the miR164e related pathway, and the previously reported RPS27AA related pathway. Knockout and overexpression of the target gene Os02g0817500 of miR5075 decreased and enhanced seed vigor, respectively. By Y2H assay, the proteins encoded by five seed vigor-related genes, Os08g0295100, Os07g0633100, REFA1, OsPER1 and OsGAPC3, were identified to interact with Os02g0817500.
Conclusions: miRNAs cooperate to regulate seed vigor in rice via an integrative gene interaction network comprising miRNA target genes and other functional genes. The result provided a basis for fully understanding the molecular mechanisms of seed vigor regulation.
Keywords: Degradome; Gene interaction network; Oryza sativa L.; Seed vigor; miRNAs.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures







Similar articles
-
Elucidation of the miR164c-Guided Gene/Protein Interaction Network Controlling Seed Vigor in Rice.Front Plant Sci. 2020 Nov 12;11:589005. doi: 10.3389/fpls.2020.589005. eCollection 2020. Front Plant Sci. 2020. PMID: 33281848 Free PMC article.
-
miR164c and miR168a regulate seed vigor in rice.J Integr Plant Biol. 2020 Apr;62(4):470-486. doi: 10.1111/jipb.12792. Epub 2019 Apr 4. J Integr Plant Biol. 2020. PMID: 30791202
-
Identification of miRNAs and Their Target Genes Associated with Sweet Corn Seed Vigor by Combined Small RNA and Degradome Sequencing.J Agric Food Chem. 2015 Jun 10;63(22):5485-91. doi: 10.1021/acs.jafc.5b00522. Epub 2015 May 29. J Agric Food Chem. 2015. PMID: 25997082
-
Rice seed storability: From molecular mechanisms to agricultural practices.Plant Sci. 2024 Nov;348:112215. doi: 10.1016/j.plantsci.2024.112215. Epub 2024 Aug 14. Plant Sci. 2024. PMID: 39151802 Review.
-
Progress in the study of functional genes related to direct seeding of rice.Mol Breed. 2023 May 24;43(6):46. doi: 10.1007/s11032-023-01388-y. eCollection 2023 Jun. Mol Breed. 2023. PMID: 37309311 Free PMC article. Review.
Cited by
-
Integrated Analysis of miRNAome and Transcriptome Identify Regulators of Elm Seed Aging.Plants (Basel). 2023 Apr 20;12(8):1719. doi: 10.3390/plants12081719. Plants (Basel). 2023. PMID: 37111942 Free PMC article.
-
Comparative miRNAome combined with transcriptome and degradome analysis reveals a novel miRNA-mRNA regulatory network associated with starch metabolism affecting pre-harvest sprouting resistance in wheat.BMC Plant Biol. 2025 Jan 24;25(1):104. doi: 10.1186/s12870-024-06039-8. BMC Plant Biol. 2025. PMID: 39856549 Free PMC article.
-
The expression of Aldo-keto reductase gene OsKOB1 coordinates ABA homeostasis and antioxidant defense to maintain seed vigor under osmotic stress.Sci Rep. 2025 Jul 28;15(1):27413. doi: 10.1038/s41598-025-13633-9. Sci Rep. 2025. PMID: 40721639 Free PMC article.
-
Identification of miRNAs Mediating Seed Storability of Maize during Germination Stage by High-Throughput Sequencing, Transcriptome and Degradome Sequencing.Int J Mol Sci. 2022 Oct 15;23(20):12339. doi: 10.3390/ijms232012339. Int J Mol Sci. 2022. PMID: 36293196 Free PMC article.
-
Correction: Degradome sequencing reveals an integrative miRNA-mediated gene interaction network regulating rice seed vigor.BMC Plant Biol. 2022 Jun 28;22(1):312. doi: 10.1186/s12870-022-03700-y. BMC Plant Biol. 2022. PMID: 35764938 Free PMC article. No abstract available.
References
-
- Perry DA. The concept of seed vigor and its relevance to seed production techniques. In: Hebblethwaite PD, editor. Seed production. London: Butterworths; 1980. pp. 585–591.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

