Anti-HIV-1 Nanobody-IgG1 Constructs With Improved Neutralization Potency and the Ability to Mediate Fc Effector Functions
- PMID: 35651621
- PMCID: PMC9150821
- DOI: 10.3389/fimmu.2022.893648
Anti-HIV-1 Nanobody-IgG1 Constructs With Improved Neutralization Potency and the Ability to Mediate Fc Effector Functions
Erratum in
-
Corrigendum: Anti-HIV-1 nanobody-IgG1 constructs with improved neutralization potency and the ability to mediate Fc effector functions.Front Immunol. 2022 Nov 22;13:1091668. doi: 10.3389/fimmu.2022.1091668. eCollection 2022. Front Immunol. 2022. PMID: 36483568 Free PMC article.
Abstract
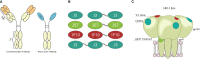
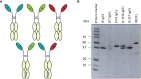
The most effective treatment for HIV-1, antiretroviral therapy, suppresses viral replication and averts the disease from progression. Nonetheless, there is a need for alternative treatments as it requires daily administration with the possibility of side effects and occurrence of drug resistance. Broadly neutralizing antibodies or nanobodies targeting the HIV-1 envelope glycoprotein are explored as alternative treatment, since they mediate viral suppression and contribute to the elimination of virus-infected cells. Besides neutralization potency and breadth, Fc-mediated effector functions of bNAbs also contribute to the in vivo efficacy. In this study multivalent J3, 2E7 and 1F10 anti-HIV-1 broadly neutralizing nanobodies were generated to improve neutralization potency and IgG1 Fc fusion was utilized to gain Fc-mediated effector functions. Bivalent and trivalent nanobodies, coupled using long glycine-serine linkers, showed increased binding to the HIV-1 Env and enhanced neutralization potency compared to the monovalent variant. Fusion of an IgG1 Fc domain to J3 improved neutralization potency compared to the J3-bihead and restored Fc-mediated effector functions such as antibody-dependent cellular phagocytosis and trogocytosis, and natural killer cell activation. Due to their neutralization breadth and potency and their ability to induce effector functions these nanobody-IgG1 constructs may prove to be valuable towards alternative HIV-1 therapies.
Keywords: Fc fusion; Fc-mediated effector functions; HIV-1; nanobodies; neutralization.
Copyright © 2022 Schriek, van Haaren, Poniman, Dekkers, Bentlage, Grobben, Vidarsson, Sanders, Verrips, Geijtenbeek, Heukers, Kootstra, de Taeye and van Gils.
Conflict of interest statement
Authors GD and RH were employed by QVQ Holding BV. Author TV was employed by VerLin BV. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




Similar articles
-
Enhancement of Antibody-Dependent Cellular Cytotoxicity and Phagocytosis in Anti-HIV-1 Human-Bovine Chimeric Broadly Neutralizing Antibodies.J Virol. 2021 Jun 10;95(13):e0021921. doi: 10.1128/JVI.00219-21. Epub 2021 Jun 10. J Virol. 2021. PMID: 33853957 Free PMC article.
-
HIV Broadly Neutralizing Antibodies Expressed as IgG3 Preserve Neutralization Potency and Show Improved Fc Effector Function.Front Immunol. 2021 Sep 10;12:733958. doi: 10.3389/fimmu.2021.733958. eCollection 2021. Front Immunol. 2021. PMID: 34566999 Free PMC article.
-
Coordinated Fc-effector and neutralization functions in HIV-infected children define a window of opportunity for HIV vaccination.AIDS. 2021 Oct 1;35(12):1895-1905. doi: 10.1097/QAD.0000000000002976. AIDS. 2021. PMID: 34115644 Free PMC article.
-
Conformational Masking and Receptor-Dependent Unmasking of Highly Conserved Env Epitopes Recognized by Non-Neutralizing Antibodies That Mediate Potent ADCC against HIV-1.Viruses. 2015 Sep 18;7(9):5115-32. doi: 10.3390/v7092856. Viruses. 2015. PMID: 26393642 Free PMC article. Review.
-
Roles of fragment crystallizable-mediated effector functions in broadly neutralizing antibody activity against HIV.Curr Opin HIV AIDS. 2020 Sep;15(5):316-323. doi: 10.1097/COH.0000000000000644. Curr Opin HIV AIDS. 2020. PMID: 32732552 Free PMC article. Review.
Cited by
-
Delivery of Fc-fusion Protein by a Recombinant Newcastle Disease Virus Vector.Appl Biochem Biotechnol. 2023 Mar;195(3):2077-2092. doi: 10.1007/s12010-022-04237-4. Epub 2022 Nov 21. Appl Biochem Biotechnol. 2023. PMID: 36417109
-
Advancing pancreatic cancer therapy by mesothelin-specific nanobody conjugation.Mol Cancer. 2025 Apr 24;24(1):124. doi: 10.1186/s12943-025-02325-7. Mol Cancer. 2025. PMID: 40275270 Free PMC article.
-
CXCR4 Is a Potential Target for Anti-HIV Gene Therapy.Int J Mol Sci. 2024 Jan 18;25(2):1187. doi: 10.3390/ijms25021187. Int J Mol Sci. 2024. PMID: 38256260 Free PMC article. Review.
-
Afucosylated broadly neutralizing antibodies enhance clearance of HIV-1 infected cells through cell-mediated killing.Commun Biol. 2024 Aug 9;7(1):964. doi: 10.1038/s42003-024-06659-8. Commun Biol. 2024. PMID: 39122901 Free PMC article.
-
Nanoscale warriors against viral invaders: a comprehensive review of Nanobodies as potential antiviral therapeutics.MAbs. 2025 Dec;17(1):2486390. doi: 10.1080/19420862.2025.2486390. Epub 2025 Apr 9. MAbs. 2025. PMID: 40201976 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

