The Case for Optimized Edge-Centric Tractography at Scale
- PMID: 35651721
- PMCID: PMC9148990
- DOI: 10.3389/fninf.2022.752471
The Case for Optimized Edge-Centric Tractography at Scale
Abstract
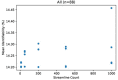
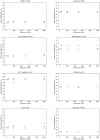
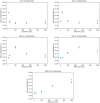
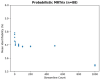
The anatomic validity of structural connectomes remains a significant uncertainty in neuroimaging. Edge-centric tractography reconstructs streamlines in bundles between each pair of cortical or subcortical regions. Although edge bundles provides a stronger anatomic embedding than traditional connectomes, calculating them for each region-pair requires exponentially greater computation. We observe that major speedup can be achieved by reducing the number of streamlines used by probabilistic tractography algorithms. To ensure this does not degrade connectome quality, we calculate the identifiability of edge-centric connectomes between test and re-test sessions as a proxy for information content. We find that running PROBTRACKX2 with as few as 1 streamline per voxel per region-pair has no significant impact on identifiability. Variation in identifiability caused by streamline count is overshadowed by variation due to subject demographics. This finding even holds true in an entirely different tractography algorithm using MRTrix. Incidentally, we observe that Jaccard similarity is more effective than Pearson correlation in calculating identifiability for our subject population.
Keywords: EDI; connectomes; diffusion MRI; edge-centric; identifiability; optimization; tractography.
Copyright © 2022 Moon, Mukherjee, Madduri, Markowitz, Cai, Palacios, Manley and Bremer.
Conflict of interest statement
GM discloses grants from the United States Department of Defense—TBI Endpoints Development Initiative (Grant #W81XWH-14-2-0176), TRACK-TBI Precision Medicine (Grant #W81XWH-18-2-0042), and TRACK-TBI NETWORK (Grant #W81XWH-15-9-0001); NIH-NINDS—TRACK-TBI (Grant #U01NS086090); and the National Football League (NFL) Scientific Advisory Board—TRACK-TBI LONGITUDINAL. The United States Department of Energy supports GM for a precision medicine collaboration. One Mind has provided funding for TRACK-TBI patients stipends and support to clinical sites. He has received an unrestricted gift from the NFL to the UCSF Foundation to support research efforts of the TRACK-TBI NETWORK. He has also received funding from NeuroTruama Sciences LLC to support TRACK-TBI data curation efforts. Additionally, Abbott Laboratories has provided funding for add-in TRACK-TBI clinical studies. AM receives funding from the Department of Defense TBI Endpoints Development Initiative (Grant #W81XWH-14-2-0176) and TRACK-TBI NETWORK (Grant #W81XWH-15-9-0001). She also receives salary support from the United States Department of Energy precision medicine collaboration and the philanthropic organization, One Mind. JM and P-TB are employed by Lawrence Livermore National Security, LLC. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
-
- Burgess G. C., Kandala S., Nolan D., Laumann T. O., Power J. D., Adeyemo B., et al. . (2016). Evaluation of denoising strategies to address motion-correlated artifacts in resting-state functional magnetic resonance imaging data from the human connectome project. Brain Connect. 6, 669–680. 10.1089/brain.2016.0435 - DOI - PMC - PubMed

