First Chromosome-Scale Assembly and Deep Floral-Bud Transcriptome of a Male Kiwifruit
- PMID: 35651931
- PMCID: PMC9149279
- DOI: 10.3389/fgene.2022.852161
First Chromosome-Scale Assembly and Deep Floral-Bud Transcriptome of a Male Kiwifruit
Keywords: PacBio and illumina sequencing; long read genome assembly; male kiwifruit genome; male—genetics; sex determining region; transcriptomic of male flower; whole-genome sequencing (WGS).
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
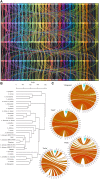
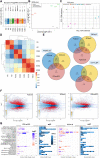
References
-
- Bushnell B. (2014). BBMap: A Fast, Accurate, Splice-Aware Aligner. United States. Available at: https://www.osti.gov/servlets/purl/1241166
LinkOut - more resources
Full Text Sources

