Uncovering BTB and CNC Homology1 (BACH1) as a Novel Cancer Therapeutic Target
- PMID: 35651942
- PMCID: PMC9149251
- DOI: 10.3389/fgene.2022.920911
Uncovering BTB and CNC Homology1 (BACH1) as a Novel Cancer Therapeutic Target
Abstract
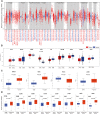
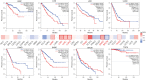
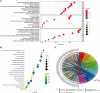
BTB and CNC homology1 (BACH1), working as a transcriptional factor, is demonstrated to function on the regulation of epigenetic modifications by complex regulatory networks. Although BACH1 is reported as an oncogene, the overall analysis of its role remains lacking. In this study, we uncovered the capacity of BACH1 as a new pan-cancer therapeutic target. We found that BACH1 is highly expressed in abundant cancers and correlated with the poor prognosis of most cancers. The mutation sites of BACH1 varied in different cancer types and correlated to patients' prognoses. The tumor mutation burden (TMB) in four cancer species and up to six tumor infiltrated immune cells had a significant relevance with BACH1. The enrichment analysis showed that the BACH1-associated genes were significantly enriched in the pathways of PD-1/L1 expression, ubiquitin-mediated proteolysis, T cell receptor, Th17 cell differentiation. We then demonstrated that BACH1 is positively correlated with the expression of many candidate genes, incluing SRPK2, GCLM, SLC40A1, and HK2 but negatively correlated with the expression of KEAP1 and GAPDH. Overall, our data shed light on BACH1's effect on latent utility in cancer targeting therapy.
Keywords: BTB and CNC homology1 (BACH1); immune cell infiltration; metabolism; therapeutic target; tumor mutation burden.
Copyright © 2022 Liu, Wang, Chen, Wu, Liao, Wang and Zhu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

