Bile Acid Conjugation on Solid Nanoparticles Enhances ASBT-Mediated Endocytosis and Chylomicron Pathway but Weakens the Transcytosis by Inducing Transport Flow in a Cellular Negative Feedback Loop
- PMID: 35652273
- PMCID: PMC9313510
- DOI: 10.1002/advs.202201414
Bile Acid Conjugation on Solid Nanoparticles Enhances ASBT-Mediated Endocytosis and Chylomicron Pathway but Weakens the Transcytosis by Inducing Transport Flow in a Cellular Negative Feedback Loop
Abstract
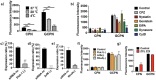
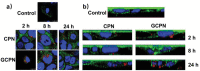
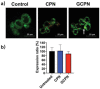
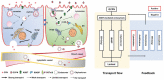
Bile acid-modified nanoparticles provide a convenient strategy to improve oral bioavailability of poorly permeable drugs by exploiting specific interactions with bile acid transporters. However, the underlying mechanisms are unknown, especially considering the different absorption sites of free bile acids (ileum) and digested fat molecules from bile acid-emulsified fat droplets (duodenum). Here, glycocholic acid (GCA)-conjugated polystyrene nanoparticles (GCPNs) are synthesized and their transport in Caco-2 cell models is studied. GCA conjugation enhances the uptake by interactions with apical sodium-dependent bile acid transporter (ASBT). A new pathway correlated with both ASBT and chylomicron pathways is identified. Meanwhile, the higher uptake of GCPNs does not lead to higher transcytosis to the same degree compared with unmodified nanoparticles (CPNs). The pharmacological and genomics study confirm that GCA conjugation changes the endocytosis mechanisms and downregulates the cellular response to the transport at gene levels, which works as a negative feedback loop and explains the higher cellular retention of GCPNs. These findings offer a solid foundation in the bile acid-based nanomedicine design, with utilizing advantages of the ASBT-mediated uptake, as well as inspiration to take comprehensive consideration of the cellular response with more developed technologies.
Keywords: apical sodium dependent bile acid transporter (ASBT); bile acid-modified nanoparticles; chylomicron pathway; transcytosis; transport feedback.
© 2022 The Authors. Advanced Science published by Wiley-VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Lennernas H., Abrahamsson B., Persson E. M., Knutson L., J. Drug Delivery Sci. Technol. 2007, 17, 237.
-
- Ganta S., Deshpande D., Korde A., Amiji M., Mol. Membr. Biol. 2010, 27, 260. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
