Tip60 acetylation of histone H3K4 temporally controls chromosome passenger complex localization
- PMID: 35653296
- PMCID: PMC9582641
- DOI: 10.1091/mbc.E21-06-0283
Tip60 acetylation of histone H3K4 temporally controls chromosome passenger complex localization
Abstract
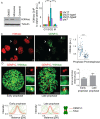
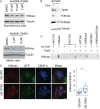
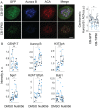
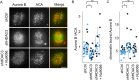
The Chromosome Passenger Complex (CPC) generates chromosome autonomous signals that regulate mitotic events critical for genome stability. Tip60 is a lysine acetyltransferase that is a tumor suppressor and is targeted for proteasomal degradation by oncogenic papilloma viruses. Mitotic regulation requires the localization of the CPC to inner centromeres, which is driven by the Haspin kinase phosphorylating histone H3 on threonine 3 (H3T3ph). Here we describe how Tip60 acetylates histone H3 at lysine 4 (H3K4ac) to block both the H3T3ph writer and the reader to ensure that this mitotic signaling cannot begin before prophase. Specifically, H3K4ac inhibits Haspin phosphorylation of H3T3 and prevents binding of the Survivin subunit to H3T3ph. Tip60 acetylates H3K4 during S/G2 at centromeres. Inhibition of Tip60 allows the CPC to bind centromeres in G2 cells, and targeting of Tip60 to centromeres prevents CPC localization in mitosis. The H3K4ac mark is removed in prophase by HDAC3 to initiate the CPC localization cascade. Together, our results suggest that Tip60 and HDAC3 temporally control H3K4 acetylation to precisely time the targeting of the CPC to inner centromeres.
Figures


Similar articles
-
Histone H3 Thr-3 phosphorylation by Haspin positions Aurora B at centromeres in mitosis.Science. 2010 Oct 8;330(6001):231-5. doi: 10.1126/science.1189435. Epub 2010 Aug 12. Science. 2010. PMID: 20705812 Free PMC article.
-
Haspin participates in AURKB recruitment to centromeres and contributes to chromosome congression in male mouse meiosis.J Cell Sci. 2022 Jul 1;135(13):jcs259546. doi: 10.1242/jcs.259546. Epub 2022 Jul 11. J Cell Sci. 2022. PMID: 35694956
-
Chromosome segregation regulation in human zygotes: altered mitotic histone phosphorylation dynamics underlying centromeric targeting of the chromosomal passenger complex.Hum Reprod. 2015 Oct;30(10):2275-91. doi: 10.1093/humrep/dev186. Epub 2015 Jul 29. Hum Reprod. 2015. PMID: 26223676
-
Haspin: a newly discovered regulator of mitotic chromosome behavior.Chromosoma. 2010 Apr;119(2):137-47. doi: 10.1007/s00412-009-0250-4. Epub 2009 Dec 8. Chromosoma. 2010. PMID: 19997740 Free PMC article. Review.
-
The chromosomal passenger complex: guiding Aurora-B through mitosis.J Cell Biol. 2006 Jun 19;173(6):833-7. doi: 10.1083/jcb.200604032. Epub 2006 Jun 12. J Cell Biol. 2006. PMID: 16769825 Free PMC article. Review.
Cited by
-
Epigenetic regulation limits competence of pluripotent stem cell-derived oocytes.EMBO J. 2023 Dec 1;42(23):e113955. doi: 10.15252/embj.2023113955. Epub 2023 Oct 18. EMBO J. 2023. PMID: 37850882 Free PMC article.
-
Non-canonical role for the BAF complex subunit DPF3 in mitosis and ciliogenesis.J Cell Sci. 2024 May 1;137(9):jcs261744. doi: 10.1242/jcs.261744. Epub 2024 May 13. J Cell Sci. 2024. PMID: 38661008 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

