Omicron BA.1 breakthrough infection drives cross-variant neutralization and memory B cell formation against conserved epitopes
- PMID: 35653438
- PMCID: PMC9162083
- DOI: 10.1126/sciimmunol.abq2427
Omicron BA.1 breakthrough infection drives cross-variant neutralization and memory B cell formation against conserved epitopes
Abstract
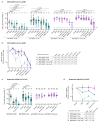
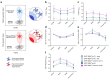
Omicron is the evolutionarily most distinct severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variant of concern (VOC) to date. We report that Omicron BA.1 breakthrough infection in BNT162b2-vaccinated individuals resulted in strong neutralizing activity against Omicron BA.1, BA.2, and previous SARS-CoV-2 VOCs but not against the Omicron sublineages BA.4 and BA.5. BA.1 breakthrough infection induced a robust recall response, primarily expanding memory B (BMEM) cells against epitopes shared broadly among variants, rather than inducing BA.1-specific B cells. The vaccination-imprinted BMEM cell pool had sufficient plasticity to be remodeled by heterologous SARS-CoV-2 spike glycoprotein exposure. Whereas selective amplification of BMEM cells recognizing shared epitopes allows for effective neutralization of most variants that evade previously established immunity, susceptibility to escape by variants that acquire alterations at hitherto conserved sites may be heightened.
Trial registration: ClinicalTrials.gov NCT04380701 NCT04949490 NCT04380701.
Figures


Comment in
-
Immune memory to SARS-CoV-2 Omicron BA.1 breakthrough infections: To change the vaccine or not?Sci Immunol. 2022 Aug 26;7(74):eabq5901. doi: 10.1126/sciimmunol.abq5901. Epub 2022 Aug 26. Sci Immunol. 2022. PMID: 35653497
References
-
- Khoury D. S., Cromer D., Reynaldi A., Schlub T. E., Wheatley A. K., Juno J. A., Subbarao K., Kent S. J., Triccas J. A., Davenport M. P., Neutralizing antibody levels are highly predictive of immune protection from symptomatic SARS-CoV-2 infection. Nat. Med. 27, 1205–1211 (2021). 10.1038/s41591-021-01377-8 - DOI - PubMed
-
- Gilbert P. B., Montefiori D. C., McDermott A. B., Fong Y., Benkeser D., Deng W., Zhou H., Houchens C. R., Martins K., Jayashankar L., Castellino F., Flach B., Lin B. C., O’Connell S., McDanal C., Eaton A., Sarzotti-Kelsoe M., Lu Y., Yu C., Borate B., van der Laan L. W. P., Hejazi N. S., Huynh C., Miller J., El Sahly H. M., Baden L. R., Baron M., De La Cruz L., Gay C., Kalams S., Kelley C. F., Andrasik M. P., Kublin J. G., Corey L., Neuzil K. M., Carpp L. N., Pajon R., Follmann D., Donis R. O., Koup R. A.; Immune Assays Team§; Moderna, Inc. Team§; Coronavirus Vaccine Prevention Network (CoVPN)/Coronavirus Efficacy (COVE) Team§; United States Government (USG)/CoVPN Biostatistics Team§ , Immune correlates analysis of the mRNA-1273 COVID-19 vaccine efficacy clinical trial. Science 375, 43–50 (2022). 10.1126/science.abm3425 - DOI - PMC - PubMed
-
- Röltgen K., Nielsen S. C. A., Silva O., Younes S. F., Zaslavsky M., Costales C., Yang F., Wirz O. F., Solis D., Hoh R. A., Wang A., Arunachalam P. S., Colburg D., Zhao S., Haraguchi E., Lee A. S., Shah M. M., Manohar M., Chang I., Gao F., Mallajosyula V., Li C., Liu J., Shoura M. J., Sindher S. B., Parsons E., Dashdorj N. J., Dashdorj N. D., Monroe R., Serrano G. E., Beach T. G., Chinthrajah R. S., Charville G. W., Wilbur J. L., Wohlstadter J. N., Davis M. M., Pulendran B., Troxell M. L., Sigal G. B., Natkunam Y., Pinsky B. A., Nadeau K. C., Boyd S. D., Immune imprinting, breadth of variant recognition, and germinal center response in human SARS-CoV-2 infection and vaccination. Cell 185, 1025–1040.e14 (2022). 10.1016/j.cell.2022.01.018 - DOI - PMC - PubMed
-
- Evans J. P., Zeng C., Carlin C., Lozanski G., Saif L. J., Oltz E. M., Gumina R. J., Liu S.-L., Neutralizing antibody responses elicited by SARS-CoV-2 mRNA vaccination wane over time and are boosted by breakthrough infection. Sci. Transl. Med. 14, eabn8057 (2022). 10.1126/scitranslmed.abn8057 - DOI - PMC - PubMed
-
- Yamayoshi S., Yasuhara A., Ito M., Akasaka O., Nakamura M., Nakachi I., Koga M., Mitamura K., Yagi K., Maeda K., Kato H., Nojima M., Pattinson D., Ogura T., Baba R., Fujita K., Nagai H., Yamamoto S., Saito M., Adachi E., Ochi J., Hattori S. I., Suzuki T., Miyazato Y., Chiba S., Okuda M., Murakami J., Hamabata T., Iwatsuki-Horimoto K., Nakajima H., Mitsuya H., Omagari N., Sugaya N., Yotsuyanagi H., Kawaoka Y., Antibody titers against SARS-CoV-2 decline, but do not disappear for several months. EClinicalMedicine 32, 100734 (2021). 10.1016/j.eclinm.2021.100734 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

