Reaction hijacking of tyrosine tRNA synthetase as a new whole-of-life-cycle antimalarial strategy
- PMID: 35653481
- PMCID: PMC7613620
- DOI: 10.1126/science.abn0611
Reaction hijacking of tyrosine tRNA synthetase as a new whole-of-life-cycle antimalarial strategy
Abstract
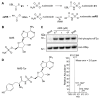
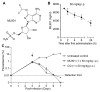
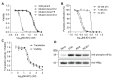
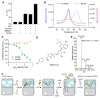
Aminoacyl transfer RNA (tRNA) synthetases (aaRSs) are attractive drug targets, and we present class I and II aaRSs as previously unrecognized targets for adenosine 5'-monophosphate-mimicking nucleoside sulfamates. The target enzyme catalyzes the formation of an inhibitory amino acid-sulfamate conjugate through a reaction-hijacking mechanism. We identified adenosine 5'-sulfamate as a broad-specificity compound that hijacks a range of aaRSs and ML901 as a specific reagent a specific reagent that hijacks a single aaRS in the malaria parasite Plasmodium falciparum, namely tyrosine RS (PfYRS). ML901 exerts whole-life-cycle-killing activity with low nanomolar potency and single-dose efficacy in a mouse model of malaria. X-ray crystallographic studies of plasmodium and human YRSs reveal differential flexibility of a loop over the catalytic site that underpins differential susceptibility to reaction hijacking by ML901.
Conflict of interest statement
S-CH, LM, M-SK, SL and AEG are (or were) employees and shareholders of Takeda. ML901 is exemplified (as compound I-27) in patent application, PCT/US2017/061094 (22).
Figures
Comment in
-
Inhibiting protein synthesis to treat malaria.Science. 2022 Jun 3;376(6597):1049-1050. doi: 10.1126/science.abq4457. Epub 2022 Jun 2. Science. 2022. PMID: 35653471
-
Thwarting protein synthesis leads to malaria parasite paralysis.Trends Parasitol. 2022 Sep;38(9):719-721. doi: 10.1016/j.pt.2022.07.001. Epub 2022 Jul 14. Trends Parasitol. 2022. PMID: 35843778
References
-
- World_Health_Organisation. World Malaria Report 2021. 2021. https://www.who.int/publications/i/item/world-malaria-report-2021 .
-
- Balikagala B, et al. Evidence of artemisinin-resistant malaria in Africa. New England Journal of Medicine. 2021;385:1163–1171. - PubMed
-
- Brownell JE, et al. Substrate-assisted inhibition of ubiquitin-like protein-activating enzymes: the NEDD8 E1 inhibitor MLN4924 forms a NEDD8-AMP mimetic in situ. Mol Cell. 2010;37:102–111. - PubMed
-
- Florini J. In: Antibiotics I: Mechanism of Action. Shaw G, editor. Springer; 1967. pp. 427–433.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

