Structural and mechanistic basis of σ-dependent transcriptional pausing
- PMID: 35653571
- PMCID: PMC9191641
- DOI: 10.1073/pnas.2201301119
Structural and mechanistic basis of σ-dependent transcriptional pausing
Abstract
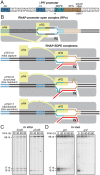
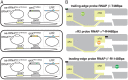
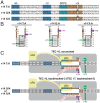
In σ-dependent transcriptional pausing, the transcription initiation factor σ, translocating with RNA polymerase (RNAP), makes sequence-specific protein–DNA interactions with a promoter-like sequence element in the transcribed region, inducing pausing. It has been proposed that, in σ-dependent pausing, the RNAP active center can access off-pathway “backtracked” states that are substrates for the transcript-cleavage factors of the Gre family and on-pathway “scrunched” states that mediate pause escape. Here, using site-specific protein–DNA photocrosslinking to define positions of the RNAP trailing and leading edges and of σ relative to DNA at the λPR′ promoter, we show directly that σ-dependent pausing in the absence of GreB in vitro predominantly involves a state backtracked by 2–4 bp, and σ-dependent pausing in the presence of GreB in vitro and in vivo predominantly involves a state scrunched by 2–3 bp. Analogous experiments with a library of 47 (∼16,000) transcribed-region sequences show that the state scrunched by 2–3 bp—and only that state—is associated with the consensus sequence, T−3N−2Y−1G+1, (where −1 corresponds to the position of the RNA 3′ end), which is identical to the consensus for pausing in initial transcription and which is related to the consensus for pausing in transcription elongation. Experiments with heteroduplex templates show that sequence information at position T−3 resides in the DNA nontemplate strand. A cryoelectron microscopy structure of a complex engaged in σ-dependent pausing reveals positions of DNA scrunching on the DNA nontemplate and template strands and suggests that position T−3 of the consensus sequence exerts its effects by facilitating scrunching.
Keywords: DNA scrunching; RNA polymerase; pausing; sigma; transcription elongation.
Conflict of interest statement
The authors declare no competing interest.
Figures



References
-
- Winkelman J. T., Nickels B. E., Ebright R. H., “The transition from transcription initiation to transcription elongation: Start-site selection, initial transcription, and promoter escape” in RNA Polymerase as a Molecular Motor, Landick R., Wang J., Strick T. R., Eds. (RSC Publishing, Cambridge, UK, ed. 2, 2021).
-
- Feklístov A., Sharon B. D., Darst S. A., Gross C. A., Bacterial sigma factors: A historical, structural, and genomic perspective. Annu. Rev. Microbiol. 68, 357–376 (2014). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

