Accurate virus identification with interpretable Raman signatures by machine learning
- PMID: 35653572
- PMCID: PMC9191668
- DOI: 10.1073/pnas.2118836119
Accurate virus identification with interpretable Raman signatures by machine learning
Abstract
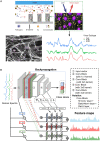
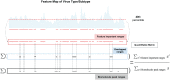
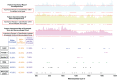
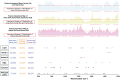
Rapid identification of newly emerging or circulating viruses is an important first step toward managing the public health response to potential outbreaks. A portable virus capture device, coupled with label-free Raman spectroscopy, holds the promise of fast detection by rapidly obtaining the Raman signature of a virus followed by a machine learning (ML) approach applied to recognize the virus based on its Raman spectrum, which is used as a fingerprint. We present such an ML approach for analyzing Raman spectra of human and avian viruses. A convolutional neural network (CNN) classifier specifically designed for spectral data achieves very high accuracy for a variety of virus type or subtype identification tasks. In particular, it achieves 99% accuracy for classifying influenza virus type A versus type B, 96% accuracy for classifying four subtypes of influenza A, 95% accuracy for differentiating enveloped and nonenveloped viruses, and 99% accuracy for differentiating avian coronavirus (infectious bronchitis virus [IBV]) from other avian viruses. Furthermore, interpretation of neural net responses in the trained CNN model using a full-gradient algorithm highlights Raman spectral ranges that are most important to virus identification. By correlating ML-selected salient Raman ranges with the signature ranges of known biomolecules and chemical functional groups—for example, amide, amino acid, and carboxylic acid—we verify that our ML model effectively recognizes the Raman signatures of proteins, lipids, and other vital functional groups present in different viruses and uses a weighted combination of these signatures to identify viruses.
Keywords: Raman spectroscopy; interpretable machine learning; virus identification.
Conflict of interest statement
The authors declare no competing interest.
Figures



References
-
- World Health Organization, Coronavirus disease 2019 (COVID-19): Situation report, 82 (2020). https://apps.who.int/iris/handle/10665/331780. Accessed 10 January 2021.
-
- Li S., et al. , Noninvasive prostate cancer screening based on serum surface-enhanced Raman spectroscopy and support vector machine. Appl. Phys. Lett. 105, 091104 (2014).
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

