Exploring Longitudinal Cough, Breath, and Voice Data for COVID-19 Progression Prediction via Sequential Deep Learning: Model Development and Validation
- PMID: 35653606
- PMCID: PMC9217153
- DOI: 10.2196/37004
Exploring Longitudinal Cough, Breath, and Voice Data for COVID-19 Progression Prediction via Sequential Deep Learning: Model Development and Validation
Abstract
Background: Recent work has shown the potential of using audio data (eg, cough, breathing, and voice) in the screening for COVID-19. However, these approaches only focus on one-off detection and detect the infection, given the current audio sample, but do not monitor disease progression in COVID-19. Limited exploration has been put forward to continuously monitor COVID-19 progression, especially recovery, through longitudinal audio data. Tracking disease progression characteristics and patterns of recovery could bring insights and lead to more timely treatment or treatment adjustment, as well as better resource management in health care systems.
Objective: The primary objective of this study is to explore the potential of longitudinal audio samples over time for COVID-19 progression prediction and, especially, recovery trend prediction using sequential deep learning techniques.
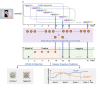
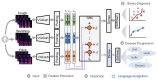
Methods: Crowdsourced respiratory audio data, including breathing, cough, and voice samples, from 212 individuals over 5-385 days were analyzed, alongside their self-reported COVID-19 test results. We developed and validated a deep learning-enabled tracking tool using gated recurrent units (GRUs) to detect COVID-19 progression by exploring the audio dynamics of the individuals' historical audio biomarkers. The investigation comprised 2 parts: (1) COVID-19 detection in terms of positive and negative (healthy) tests using sequential audio signals, which was primarily assessed in terms of the area under the receiver operating characteristic curve (AUROC), sensitivity, and specificity, with 95% CIs, and (2) longitudinal disease progression prediction over time in terms of probability of positive tests, which was evaluated using the correlation between the predicted probability trajectory and self-reported labels.
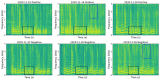
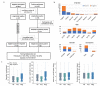
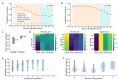
Results: We first explored the benefits of capturing longitudinal dynamics of audio biomarkers for COVID-19 detection. The strong performance, yielding an AUROC of 0.79, a sensitivity of 0.75, and a specificity of 0.71 supported the effectiveness of the approach compared to methods that do not leverage longitudinal dynamics. We further examined the predicted disease progression trajectory, which displayed high consistency with longitudinal test results with a correlation of 0.75 in the test cohort and 0.86 in a subset of the test cohort with 12 (57.1%) of 21 COVID-19-positive participants who reported disease recovery. Our findings suggest that monitoring COVID-19 evolution via longitudinal audio data has potential in the tracking of individuals' disease progression and recovery.
Conclusions: An audio-based COVID-19 progression monitoring system was developed using deep learning techniques, with strong performance showing high consistency between the predicted trajectory and the test results over time, especially for recovery trend predictions. This has good potential in the postpeak and postpandemic era that can help guide medical treatment and optimize hospital resource allocations. The changes in longitudinal audio samples, referred to as audio dynamics, are associated with COVID-19 progression; thus, modeling the audio dynamics can potentially capture the underlying disease progression process and further aid COVID-19 progression prediction. This framework provides a flexible, affordable, and timely tool for COVID-19 tracking, and more importantly, it also provides a proof of concept of how telemonitoring could be applicable to respiratory diseases monitoring, in general.
Keywords: COVID-19; COVID-19 progression; audio; deep learning; longitudinal study; mobile health.
©Ting Dang, Jing Han, Tong Xia, Dimitris Spathis, Erika Bondareva, Chloë Siegele-Brown, Jagmohan Chauhan, Andreas Grammenos, Apinan Hasthanasombat, R Andres Floto, Pietro Cicuta, Cecilia Mascolo. Originally published in the Journal of Medical Internet Research (https://www.jmir.org), 21.06.2022.
Conflict of interest statement
Conflicts of Interest: None declared.
Figures


References
-
- Vogels CBF, Brito AF, Wyllie AL, Fauver JR, Ott IM, Kalinich CC, Petrone ME, Casanovas-Massana A, Catherine Muenker M, Moore AJ, Klein J, Lu P, Lu-Culligan A, Jiang X, Kim DJ, Kudo E, Mao T, Moriyama M, Oh JE, Park A, Silva J, Song E, Takahashi T, Taura M, Tokuyama M, Venkataraman A, Weizman O-E, Wong P, Yang Y, Cheemarla NR, White EB, Lapidus S, Earnest R, Geng B, Vijayakumar P, Odio C, Fournier J, Bermejo S, Farhadian S, Dela Cruz CS, Iwasaki A, Ko AI, Landry ML, Foxman EF, Grubaugh ND. Analytical sensitivity and efficiency comparisons of SARS-CoV-2 RT-qPCR primer-probe sets. Nat Microbiol. 2020 Oct;5(10):1299–1305. doi: 10.1038/s41564-020-0761-6.10.1038/s41564-020-0761-6 - DOI - PMC - PubMed
-
- Ates HC, Yetisen AK, Güder F, Dincer C. Wearable devices for the detection of COVID-19. Nat Electron. 2021 Jan 25;4(1):13–14. doi: 10.1038/s41928-020-00533-1. - DOI
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

