Thiamine metabolism genes in diatoms are not regulated by thiamine despite the presence of predicted riboswitches
- PMID: 35653609
- PMCID: PMC9544697
- DOI: 10.1111/nph.18296
Thiamine metabolism genes in diatoms are not regulated by thiamine despite the presence of predicted riboswitches
Abstract
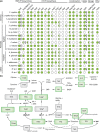
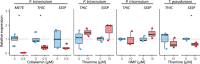
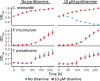
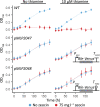
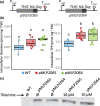
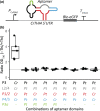
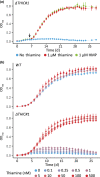
Thiamine pyrophosphate (TPP), an essential co-factor for all species, is biosynthesised through a metabolically expensive pathway regulated by TPP riboswitches in bacteria, fungi, plants and green algae. Diatoms are microalgae responsible for c. 20% of global primary production. They have been predicted to contain TPP aptamers in the 3'UTR of some thiamine metabolism-related genes, but little information is known about their function and regulation. We used bioinformatics, antimetabolite growth assays, RT-qPCR, targeted mutagenesis and reporter constructs to test whether the predicted TPP riboswitches respond to thiamine supplementation in diatoms. Gene editing was used to investigate the functions of the genes with associated TPP riboswitches in Phaeodactylum tricornutum. We found that thiamine-related genes with putative TPP aptamers are not responsive to supplementation with thiamine or its precursor 4-amino-5-hydroxymethyl-2-methylpyrimidine (HMP), and targeted mutation of the TPP aptamer in the THIC gene encoding HMP-P synthase does not deregulate thiamine biosynthesis in P. tricornutum. Through genome editing we established that PtTHIC is essential for thiamine biosynthesis and another gene, PtSSSP, is necessary for thiamine uptake. Our results highlight the importance of experimentally testing bioinformatic aptamer predictions and provide new insights into the thiamine metabolism shaping the structure of marine microbial communities with global biogeochemical importance.
Keywords: Phaeodactylum tricornutum; CRISPR/Cas9; TPP riboswitch; aptamer prediction; diatoms; thiamine biosynthesis; thiamine uptake.
© 2022 The Authors. New Phytologist © 2022 New Phytologist Foundation.
Figures


References
-
- Beilharz TH, Preiss T. 2009. Transcriptome‐wide measurement of mRNA polyadenylation state. Methods 48: 294–300. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- BB/L002957/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/M011194/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/M018180/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/R021694/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources

