Analysis of immune related gene expression profiles and immune cell components in patients with Barrett esophagus
- PMID: 35654816
- PMCID: PMC9163054
- DOI: 10.1038/s41598-022-13200-6
Analysis of immune related gene expression profiles and immune cell components in patients with Barrett esophagus
Abstract
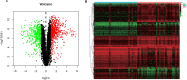
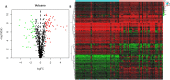
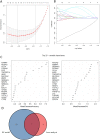
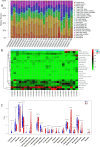
Barrett's esophagus (BE) is a well-known precancerous condition of esophageal adenocarcinoma. However, the immune cells and immune related genes involved in BE development and progression are not fully understood. Therefore, our study attempted to investigate the roles of immune cells and immune related genes in BE patients. The raw gene expression data were downloaded from the GEO database. The limma package in R was used to screen differentially expressed genes (DEGs). Then we performed the least absolute shrinkage and selection operator (LASSO) and random forest (RF) analyses to screen key genes. The proportion of infiltrated immune cells was evaluated using the CIBERSORT algorithm between BE and normal esophagus (NE) samples. The spearman index was used to show the correlations of immune genes and immune cells. Receiver operating characteristic (ROC) curves were used to assess the diagnostic value of key genes in BE. A total of 103 differentially expressed immune-related genes were identified between BE samples and normal samples. Then, 7 genes (CD1A, LTF, FABP4, PGC, TCF7L2, INSR,SEMA3C) were obtained after Lasso analysis and RF modeling. CIBERSORT analysis revealed that resting CD4 T memory cells and gamma delta T cells were present at significantly lower levels in BE samples. Moreover, plasma cell and regulatory T cells were present at significantly higher levels in BE samples than in NE samples. INSR had the highest AUC values in ROC analysis. We identified 7 immune related genes and 4 different immune cells in our study, that may play vital roles in the occurrence and development of BE. Our findings improve the understanding of the molecular mechanisms of BE.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
Biomarker identification and trans-regulatory network analyses in esophageal adenocarcinoma and Barrett's esophagus.World J Gastroenterol. 2019 Jan 14;25(2):233-244. doi: 10.3748/wjg.v25.i2.233. World J Gastroenterol. 2019. PMID: 30670912 Free PMC article.
-
Analysis of immune cell components and immune-related gene expression profiles in peripheral blood of patients with type 1 diabetes mellitus.J Transl Med. 2021 Jul 26;19(1):319. doi: 10.1186/s12967-021-02991-3. J Transl Med. 2021. PMID: 34311758 Free PMC article.
-
Identification of miRNAs and genes for predicting Barrett's esophagus progressing to esophageal adenocarcinoma using miRNA-mRNA integrated analysis.PLoS One. 2021 Nov 24;16(11):e0260353. doi: 10.1371/journal.pone.0260353. eCollection 2021. PLoS One. 2021. PMID: 34818353 Free PMC article.
-
Role of epigenetic alterations in the pathogenesis of Barrett's esophagus and esophageal adenocarcinoma.Int J Clin Exp Pathol. 2012;5(5):382-96. Epub 2012 May 23. Int J Clin Exp Pathol. 2012. PMID: 22808291 Free PMC article. Review.
-
Predictors of Progression to High-Grade Dysplasia or Adenocarcinoma in Barrett's Esophagus.Gastroenterol Clin North Am. 2015 Jun;44(2):299-315. doi: 10.1016/j.gtc.2015.02.005. Epub 2015 Mar 31. Gastroenterol Clin North Am. 2015. PMID: 26021196 Free PMC article. Review.
Cited by
-
Association between systemic immune-inflammation index and low muscle mass in US adults: a cross-sectional study.BMC Public Health. 2023 Jul 24;23(1):1416. doi: 10.1186/s12889-023-16338-8. BMC Public Health. 2023. PMID: 37488531 Free PMC article.
-
Lactate dehydrogenase to albumin ratio is associated with in-hospital mortality in patients with acute heart failure: Data from the MIMIC-III database.Open Med (Wars). 2024 Mar 19;19(1):20240901. doi: 10.1515/med-2024-0901. eCollection 2024. Open Med (Wars). 2024. PMID: 38584822 Free PMC article.
-
Association of Dietary Inflammation Index and Helicobacter pylori Immunoglobulin G Seropositivity in US Adults: A Population-Based Study.Mediators Inflamm. 2023 Jul 28;2023:8880428. doi: 10.1155/2023/8880428. eCollection 2023. Mediators Inflamm. 2023. PMID: 37545737 Free PMC article.
-
Identification of PGC as a Potential Biomarker for Progression from Barrett's Esophagus to Esophageal Adenocarcinoma: A Comprehensive Bioinformatic Analysis.Diagnostics (Basel). 2024 Dec 19;14(24):2863. doi: 10.3390/diagnostics14242863. Diagnostics (Basel). 2024. PMID: 39767224 Free PMC article.
References
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

