A stromal Integrated Stress Response activates perivascular cancer-associated fibroblasts to drive angiogenesis and tumour progression
- PMID: 35654839
- PMCID: PMC9203279
- DOI: 10.1038/s41556-022-00918-8
A stromal Integrated Stress Response activates perivascular cancer-associated fibroblasts to drive angiogenesis and tumour progression
Abstract
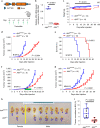
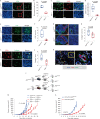
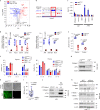
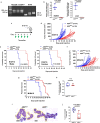
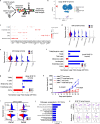
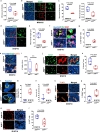
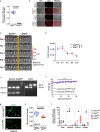
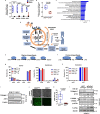
Bidirectional signalling between the tumour and stroma shapes tumour aggressiveness and metastasis. ATF4 is a major effector of the Integrated Stress Response, a homeostatic mechanism that couples cell growth and survival to bioenergetic demands. Using conditional knockout ATF4 mice, we show that global, or fibroblast-specific loss of host ATF4, results in deficient vascularization and a pronounced growth delay of syngeneic melanoma and pancreatic tumours. Single-cell transcriptomics of tumours grown in Atf4Δ/Δ mice uncovered a reduction in activation markers in perivascular cancer-associated fibroblasts (CAFs). Atf4Δ/Δ fibroblasts displayed significant defects in collagen biosynthesis and deposition and a reduced ability to support angiogenesis. Mechanistically, ATF4 regulates the expression of the Col1a1 gene and levels of glycine and proline, the major amino acids of collagen. Analyses of human melanoma and pancreatic tumours revealed a strong correlation between ATF4 and collagen levels. Our findings establish stromal ATF4 as a key driver of CAF functionality, malignant progression and metastasis.
© 2022. The Author(s).
Conflict of interest statement
C.K. is the scientific founder and holds equity position in Veltion Therapeutics. The remaining authors declare no competing interests.
Figures








Comment in
-
Stress response regulates cancer fibroblasts.Nat Cell Biol. 2022 Jun;24(6):812-814. doi: 10.1038/s41556-022-00930-y. Nat Cell Biol. 2022. PMID: 35654838 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

