Targeted isolation of diverse human protective broadly neutralizing antibodies against SARS-like viruses
- PMID: 35654851
- PMCID: PMC10083051
- DOI: 10.1038/s41590-022-01222-1
Targeted isolation of diverse human protective broadly neutralizing antibodies against SARS-like viruses
Abstract
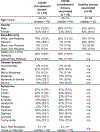
The emergence of current severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variants of concern (VOCs) and potential future spillovers of SARS-like coronaviruses into humans pose a major threat to human health and the global economy. Development of broadly effective coronavirus vaccines that can mitigate these threats is needed. Here, we utilized a targeted donor selection strategy to isolate a large panel of human broadly neutralizing antibodies (bnAbs) to sarbecoviruses. Many of these bnAbs are remarkably effective in neutralizing a diversity of sarbecoviruses and against most SARS-CoV-2 VOCs, including the Omicron variant. Neutralization breadth is achieved by bnAb binding to epitopes on a relatively conserved face of the receptor-binding domain (RBD). Consistent with targeting of conserved sites, select RBD bnAbs exhibited protective efficacy against diverse SARS-like coronaviruses in a prophylaxis challenge model in vivo. These bnAbs provide new opportunities and choices for next-generation antibody prophylactic and therapeutic applications and provide a molecular basis for effective design of pan-sarbecovirus vaccines.
© 2022. The Author(s), under exclusive licence to Springer Nature America, Inc.
Conflict of interest statement
Competing interests: W.H., R.M., G.S., K.D., T.F.R., D.R.B. and R.A. are listed as inventors on pending patent applications describing the sarbecovirus broadly neutralizing antibodies isolated in this study. A.B.W, I.A.W and D.R.B. receive research funding from Adagio. RSB and LEG have ongoing collaborations with Adagio. All other authors have no competing interests to declare.
Figures


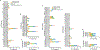





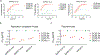






Update of
-
Targeted isolation of panels of diverse human protective broadly neutralizing antibodies against SARS-like viruses.bioRxiv [Preprint]. 2022 Feb 8:2021.09.08.459480. doi: 10.1101/2021.09.08.459480. bioRxiv. 2022. Update in: Nat Immunol. 2022 Jun;23(6):960-970. doi: 10.1038/s41590-022-01222-1. PMID: 35169804 Free PMC article. Updated. Preprint.
Comment in
-
Catching the breadth of broadly protective antibodies to SARS-CoV-2.Nat Immunol. 2022 Jun;23(6):828-829. doi: 10.1038/s41590-022-01225-y. Nat Immunol. 2022. PMID: 35654852 No abstract available.
References
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

