Focal Adhesion Kinase (FAK)-Hippo/YAP transduction signaling mediates the stimulatory effects exerted by S100A8/A9-RAGE system in triple-negative breast cancer (TNBC)
- PMID: 35655319
- PMCID: PMC9164429
- DOI: 10.1186/s13046-022-02396-0
Focal Adhesion Kinase (FAK)-Hippo/YAP transduction signaling mediates the stimulatory effects exerted by S100A8/A9-RAGE system in triple-negative breast cancer (TNBC)
Abstract
Background: Understanding the intricate signaling network involved in triple-negative breast cancer (TNBC) represents a challenge for developing novel therapeutic approaches. Here, we aim to provide novel mechanistic insights on the function of the S100A8/A9-RAGE system in TNBC.
Methods: TNM plot analyzer, Kaplan-Meier plotter, Meta-analysis, GEPIA2 and GOBO publicly available datasets were used to evaluate the clinical significance of S100A8/A9 and expression levels of S100A8/A9, RAGE and Filamin family members in breast cancer (BC) subtypes. METABRIC database and Cox proportional hazard model defined the clinical impact of high RAGE expression in BC patients. Multiple bioinformatics programs identified the main enriched pathways within high RAGE expression BC cohorts. By lentiviral system, TNBC cells were engineered to overexpress RAGE. Western blotting, immunofluorescence, nucleus/cytoplasm fractionation, qRT-PCR, gene silencing and luciferase experiments were performed to identify signal transduction mediators engaged by RAGE upon stimulation with S100A8/A9 in TNBC cells. Proliferation, colony formation and transwell migration assays were carried out to evaluate the growth and migratory capacity of TNBC cells. Statistical analysis was performed by ANOVA and independent t-tests.
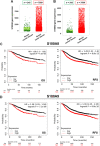
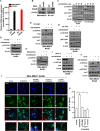
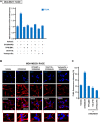
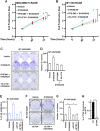
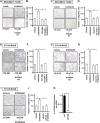
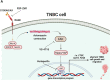
Results: We found a remarkable high expression of S100A8 and S100A9 in BC, particularly in HER2-positive and TNBC, with the latter associated to worst clinical outcomes. In addition, high RAGE expression correlated with a poor overall survival in BC. Next, we determined that the S100A8/A9-RAGE system triggers FAK activation by engaging a cytoskeleton mechanosensing complex in TNBC cells. Through bioinformatics analysis, we identified the Hippo pathway as the most enriched in BC patients expressing high RAGE levels. In accordance with these data, we demonstrated the involvement of S100A8/A9-RAGE-FAK signaling in the control of Hippo/YAP activities, and we established the crucial contribution of RAGE-FAK-YAP circuitry in the growth and migratory effects initiated by S100A8/A9 in TNBC cells.
Conclusions: The present study provides novel mechanistic insights on RAGE actions in TNBC. Moreover, our findings suggest that RAGE-FAK-YAP transduction pathway could be exploited as a druggable system halting the aggressive TNBC subtype.
Keywords: BT-549; FAK; Hippo/YAP; MDA-MB-231; RAGE; S100A8/A9; TNBC.
© 2022. The Author(s).
Conflict of interest statement
The authors disclosed no conflicts of interest. Nijiro Nohata is an employee of MSD K.K., a subsidiary of Merck & Co., Inc., and reports personal fees from MSD K. K outside of this study.
Figures



Similar articles
-
Focal adhesion kinase (FAK) activation by estrogens involves GPER in triple-negative breast cancer cells.J Exp Clin Cancer Res. 2019 Feb 6;38(1):58. doi: 10.1186/s13046-019-1056-8. J Exp Clin Cancer Res. 2019. PMID: 30728047 Free PMC article.
-
IGF-1/IGF-1R/FAK/YAP Transduction Signaling Prompts Growth Effects in Triple-Negative Breast Cancer (TNBC) Cells.Cells. 2020 Apr 18;9(4):1010. doi: 10.3390/cells9041010. Cells. 2020. PMID: 32325700 Free PMC article.
-
ZNF213 negatively controls triple negative breast cancer progression via Hippo/YAP signaling.Cancer Sci. 2021 Jul;112(7):2714-2727. doi: 10.1111/cas.14916. Epub 2021 May 3. Cancer Sci. 2021. PMID: 33939216 Free PMC article.
-
Calprotectin and the Initiation and Progression of Head and Neck Cancer.J Dent Res. 2018 Jun;97(6):674-682. doi: 10.1177/0022034518756330. Epub 2018 Feb 14. J Dent Res. 2018. PMID: 29443623 Free PMC article. Review.
-
Hippo signaling pathway: A comprehensive gene expression profile analysis in breast cancer.Biomed Pharmacother. 2022 Jul;151:113144. doi: 10.1016/j.biopha.2022.113144. Epub 2022 May 25. Biomed Pharmacother. 2022. PMID: 35623167 Review.
Cited by
-
S100A8/A9 as a risk factor for breast cancer negatively regulated by DACH1.Biomark Res. 2023 Dec 13;11(1):106. doi: 10.1186/s40364-023-00548-8. Biomark Res. 2023. PMID: 38093319 Free PMC article.
-
RNF31 represses cell progression and immune evasion via YAP/PD-L1 suppression in triple negative breast Cancer.J Exp Clin Cancer Res. 2022 Dec 29;41(1):364. doi: 10.1186/s13046-022-02576-y. J Exp Clin Cancer Res. 2022. PMID: 36581998 Free PMC article.
-
Ca2+ signaling and the Hippo pathway: Intersections in cellular regulation.Cell Signal. 2023 Oct;110:110846. doi: 10.1016/j.cellsig.2023.110846. Epub 2023 Aug 6. Cell Signal. 2023. PMID: 37549859 Free PMC article. Review.
-
The Ephrin tyrosine kinase a3 (EphA3) is a novel mediator of RAGE-prompted motility of breast cancer cells.J Exp Clin Cancer Res. 2023 Jul 12;42(1):164. doi: 10.1186/s13046-023-02747-5. J Exp Clin Cancer Res. 2023. PMID: 37434266 Free PMC article.
-
Enhancing radiotherapy in triple-negative breast cancer with hesperetin-induced ferroptosis via AURKA targeting nanocomposites.J Nanobiotechnology. 2024 Nov 30;22(1):744. doi: 10.1186/s12951-024-02987-3. J Nanobiotechnology. 2024. PMID: 39614277 Free PMC article.
References
-
- Mittendorf EA, Zhang H, Barrios CH, Saji S, Jung KH, Hegget R, et al. Neoadjuvant atezolizumab in combination with sequential nab-paclitaxel and anthracycline-based chemotherapy versus placebo and chemotherapy in patients with early-stage triple-negative breast cancer (IMpassion031): a randomised, double-blind, phase 3 trial. Lancet. 2020;396(10257):1090–1100. doi: 10.1016/S0140-6736(20)31953-X. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

