m6A Regulator-Mediated Methylation Modification Patterns and Characteristics in COVID-19 Patients
- PMID: 35655464
- PMCID: PMC9152098
- DOI: 10.3389/fpubh.2022.914193
m6A Regulator-Mediated Methylation Modification Patterns and Characteristics in COVID-19 Patients
Abstract
Background: RNA N6-methyladenosine (m6A) regulators may be necessary for diverse viral infectious diseases, and serve pivotal roles in various physiological functions. However, the potential roles of m6A regulators in coronavirus disease 2019 (COVID-19) remain unclear.
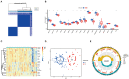
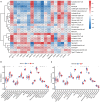
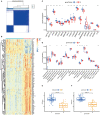
Methods: The gene expression profile of patients with or without COVID-19 was acquired from Gene Expression Omnibus (GEO) database, and bioinformatics analysis of differentially expressed genes was conducted. Random forest modal and nomogram were established to predict the occurrence of COVID-19. Afterward, the consensus clustering method was utilized to establish two different m6A subtypes, and associations between subtypes and immunity were explored.
Results: Based on the transcriptional data from GSE157103, we observed that the m6A modification level was markedly enriched in the COVID-19 patients than those in the non-COVID-19 patients. And 18 essential m6A regulators were identified with differential analysis between patients with or without COVID-19. The random forest model was utilized to determine 8 optimal m6A regulators for predicting the emergence of COVID-19. We then established a nomogram based on these regulators, and its predictive reliability was validated by decision curve analysis. The consensus clustering algorithm was conducted to categorize COVID-19 patients into two m6A subtypes from the identified m6A regulators. The patients in cluster A were correlated with activated T-cell functions and may have a superior prognosis.
Conclusions: Collectively, m6A regulators may be involved in the prevalence of COVID-19 patients. Our exploration of m6A subtypes may benefit the development of subsequent treatment modalities for COVID-19.
Keywords: COVID-19; consensus clustering; diagnostic biomarkers; m6A methylation modification; m6A regulators.
Copyright © 2022 Qing, Chen and Wang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

