Metal binding and interdomain thermodynamics of mammalian metallothionein-3: enthalpically favoured Cu+ supplants entropically favoured Zn2+ to form Cu4 + clusters under physiological conditions
- PMID: 35655557
- PMCID: PMC9093145
- DOI: 10.1039/d2sc00676f
Metal binding and interdomain thermodynamics of mammalian metallothionein-3: enthalpically favoured Cu+ supplants entropically favoured Zn2+ to form Cu4 + clusters under physiological conditions
Abstract
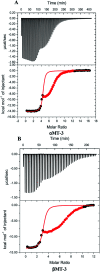
Metallothioneins (MTs) are a ubiquitous class of small metal-binding proteins involved in metal homeostasis and detoxification. While known for their high affinity for d10 metal ions, there is a surprising dearth of thermodynamic data on metals binding to MTs. In this study, Zn2+ and Cu+ binding to mammalian metallothionein-3 (MT-3) were quantified at pH 7.4 by isothermal titration calorimetry (ITC). Zn2+ binding was measured by chelation titrations of Zn7MT-3, while Cu+ binding was measured by Zn2+ displacement from Zn7MT-3 with competition from glutathione (GSH). Titrations in multiple buffers enabled a detailed analysis that yielded condition-independent values for the association constant (K) and the change in enthalpy (ΔH) and entropy (ΔS) for these metal ions binding to MT-3. Zn2+ was also chelated from the individual α and β domains of MT-3 to quantify the thermodynamics of inter-domain interactions in metal binding. Comparative titrations of Zn7MT-2 with Cu+ revealed that both MT isoforms have similar Cu+ affinities and binding thermodynamics, indicating that ΔH and ΔS are determined primarily by the conserved Cys residues. Inductively coupled plasma mass spectrometry (ICP-MS) analysis and low temperature luminescence measurements of Cu-replete samples showed that both proteins form two Cu4 +-thiolate clusters when Cu+ displaces Zn2+ under physiological conditions. Comparison of the Zn2+ and Cu+ binding thermodynamics reveal that enthalpically-favoured Cu+, which forms Cu4 +-thiolate clusters, displaces the entropically-favoured Zn2+. These results provide a detailed thermodynamic analysis of d10 metal binding to these thiolate-rich proteins and quantitative support for, as well as molecular insight into, the role that MT-3 plays in the neuronal chemistry of copper.
This journal is © The Royal Society of Chemistry.
Conflict of interest statement
There are no conflicts to declare.
Figures



Similar articles
-
Thermodynamic origin of the affinity, selectivity, and domain specificity of metallothionein for essential and toxic metal ions.Metallomics. 2024 Oct 4;16(10):mfae041. doi: 10.1093/mtomcs/mfae041. Metallomics. 2024. PMID: 39289027
-
Thermodynamics of Pb(ii) and Zn(ii) binding to MT-3, a neurologically important metallothionein.Metallomics. 2016 Jun 1;8(6):605-17. doi: 10.1039/c5mt00209e. Metallomics. 2016. PMID: 26757944
-
Non-coordinative metal selectivity bias in human metallothioneins metal-thiolate clusters.Metallomics. 2018 Dec 12;10(12):1777-1791. doi: 10.1039/c8mt00264a. Metallomics. 2018. PMID: 30420986 Free PMC article.
-
Unravelling the mechanistic details of metal binding to mammalian metallothioneins from stoichiometric, kinetic, and binding affinity data.Dalton Trans. 2018 Mar 12;47(11):3613-3637. doi: 10.1039/c7dt03319b. Dalton Trans. 2018. PMID: 29431781 Review.
-
Challenging conventional wisdom: single domain metallothioneins.Metallomics. 2014 Apr;6(4):702-28. doi: 10.1039/c3mt00216k. Metallomics. 2014. PMID: 24469686 Review.
Cited by
-
Oxidative pathways of apo, partially, and fully Zn(II)- and Cd(II)-metalated human metallothionein-3 are dominated by disulfide bond formation.FEBS J. 2025 Feb;292(3):619-634. doi: 10.1111/febs.17333. Epub 2024 Dec 1. FEBS J. 2025. PMID: 39617990 Free PMC article.
-
CUP1 Metallothionein from Healthy Saccharomyces cerevisiae Colocalizes to the Cytosol and Mitochondrial Intermembrane Space.Biochemistry. 2023 Jan 3;62(1):62-74. doi: 10.1021/acs.biochem.2c00481. Epub 2022 Dec 12. Biochemistry. 2023. PMID: 36503220 Free PMC article.
-
Ion mobility mass spectrometry and molecular dynamics simulations unravel the conformational stability of zinc metallothionein-2 species.Chem Commun (Camb). 2023 Apr 11;59(30):4471-4474. doi: 10.1039/d2cc06559b. Chem Commun (Camb). 2023. PMID: 36960761 Free PMC article.
-
63Cu(I) binding to human kidney 68Zn7-βα MT1A: determination of Cu(I)-thiolate cluster domain specificity from ESI-MS and room temperature phosphorescence spectroscopy.Metallomics. 2023 Jan 10;15(1):mfac101. doi: 10.1093/mtomcs/mfac101. Metallomics. 2023. PMID: 36583699 Free PMC article.
-
Important Structural Features of Thiolate-Rich Four-Helix Bundles for Cu(I) Uptake and Removal.Inorg Chem. 2023 May 1;62(17):6617-6628. doi: 10.1021/acs.inorgchem.2c04490. Epub 2023 Apr 14. Inorg Chem. 2023. PMID: 37057906 Free PMC article.
References
-
- Capdevila M. Bofill R. Palacios O. Atrian S. State-of-the-art of metallothioneins at the beginning of the 21st century. Coord. Chem. Rev. 2012;256:46–62. doi: 10.1016/j.ccr.2011.07.006. - DOI
-
- Hartmann H.-J. and Weser U., Metallothioneins, in Biological Inorganic Chemistry, ed. I. Bertini, H. B. Gray, E. I. Stiefel, and J. S. Valentine, University Science Books, Sausalito, 2007, pp. 156–174
Grants and funding
LinkOut - more resources
Full Text Sources

