Modeling long-range dependencies for weakly supervised disease classification and localization on chest X-ray
- PMID: 35655823
- PMCID: PMC9131331
- DOI: 10.21037/qims-21-1117
Modeling long-range dependencies for weakly supervised disease classification and localization on chest X-ray
Abstract
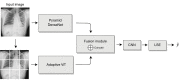
Background: Computer-aided diagnosis based on chest X-ray (CXR) is an exponentially growing field of research owing to the development of deep learning, especially convolutional neural networks (CNNs). However, due to the intrinsic locality of convolution operations, CNNs cannot model long-range dependencies. Although vision transformers (ViTs) have recently been proposed to alleviate this limitation, those trained on patches cannot learn any dependencies for inter-patch pixels and thus, are insufficient for medical image detection. To address this problem, in this paper, we propose a CXR detection method which integrates CNN with a ViT for modeling patch-wise and inter-patch dependencies.
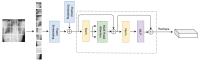
Methods: We experimented on the ChestX-ray14 dataset and followed the official training-test set split. Because the training data only had global annotations, the detection network was weakly supervised. A DenseNet with a feature pyramid structure was designed and integrated with an adaptive ViT to model inter-patch and patch-wise long-range dependencies and obtain fine-grained feature maps. We compared the performance using our method with that of other disease detection methods.
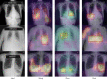
Results: For disease classification, our method achieved the best result among all the disease detection methods, with a mean area under the curve (AUC) of 0.829. For lesion localization, our method achieved significantly higher intersection of the union (IoU) scores on the test images with bounding box annotations than did the other detection methods. The visualized results showed that our predictions were more accurate and detailed. Furthermore, evaluation of our method in an external validation dataset demonstrated its generalization ability.
Conclusions: Our proposed method achieves the new state of the art for thoracic disease classification and weakly supervised localization. It has potential to assist in clinical decision-making.
Keywords: Long-range dependencies; chest X-rays (CXRs); disease classification; localization; vision transformer (ViT).
2022 Quantitative Imaging in Medicine and Surgery. All rights reserved.
Conflict of interest statement
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://qims.amegroups.com/article/view/10.21037/qims-21-1117/coif). FL has a pending patent (No. 202110640995.8). The other authors have no conflicts of interest to declare.
Figures




References
-
- Yan C, Yao J, Li R, Xu Z, Huang J. Weakly Supervised Deep Learning for Thoracic Disease Classification and Localization on Chest X-rays. Proceedings of the 2018 ACM International Conference on Bioinformatics, Computational Biology, and Health Informatics; Washington, DC, USA: Association for Computing Machinery; 2018:103-4.
LinkOut - more resources
Full Text Sources
