The Old Yellow Enzyme OfrA Fosters Staphylococcus aureus Survival via Affecting Thiol-Dependent Redox Homeostasis
- PMID: 35656003
- PMCID: PMC9152700
- DOI: 10.3389/fmicb.2022.888140
The Old Yellow Enzyme OfrA Fosters Staphylococcus aureus Survival via Affecting Thiol-Dependent Redox Homeostasis
Abstract
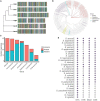
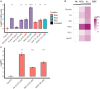
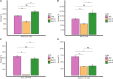
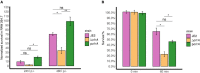
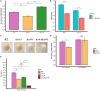
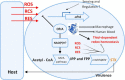
Old yellow enzymes (OYEs) are widely found in the bacterial, fungal, and plant kingdoms but absent in humans and have been used as biocatalysts for decades. However, OYEs' physiological function in bacterial stress response and infection situations remained enigmatic. As a pathogen, the Gram-positive bacterium Staphylococcus aureus adapts to numerous stress conditions during pathogenesis. Here, we show that in S. aureus genome, two paralogous genes (ofrA and ofrB) encode for two OYEs. We conducted a bioinformatic analysis and found that ofrA is conserved among all publicly available representative staphylococcal genomes and some Firmicutes. Expression of ofrA is induced by electrophilic, oxidative, and hypochlorite stress in S. aureus. Furthermore, ofrA contributes to S. aureus survival against reactive electrophilic, oxygen, and chlorine species (RES, ROS, and RCS) via thiol-dependent redox homeostasis. At the host-pathogen interface, S. aureusΔofrA has defective survival in macrophages and whole human blood and decreased staphyloxanthin production. Overall, our results shed the light onto a novel stress response strategy in the important human pathogen S. aureus.
Keywords: MRSA; ROS; blood; electrophilic stress; phagocytes; quinone; stress response.
Copyright © 2022 Ibrahim and Ohlsen.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

