Autophagy and Renal Fibrosis
- PMID: 35656109
- PMCID: PMC9116923
- DOI: 10.14336/AD.2021.1027
Autophagy and Renal Fibrosis
Abstract
Renal fibrosis is a common process of almost all the chronic kidney diseases progressing to end-stage kidney disease. As a highly conserved lysosomal protein degradation pathway, autophagy is responsible for degrading protein aggregates, damaged organelles, or invading pathogens to maintain intracellular homeostasis. Growing evidence reveals that autophagy is involved in the progression of renal fibrosis, both in the tubulointerstitial compartment and in the glomeruli. Nevertheless, the specific role of autophagy in renal fibrosis has still not been fully understood. Therefore, in this review we will describe the characteristics of autophagy and summarize the recent advances in understanding the functions of autophagy in renal fibrosis. Moreover, the problem existing in this field and the possibility of autophagy as the potential therapeutic target for renal fibrosis have also been discussed.
Keywords: autophagy; cellular senescence; chronic kidney disease; renal fibrosis; senescence-associated secretory phenotype.
Copyright: © 2022 Liang et al.
Conflict of interest statement
Competing interests All authors declare no conflict of interest.
Figures
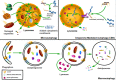
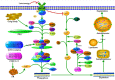
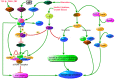
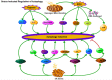
References
-
- Keith DS, Nichols GA, Gullion CM, Brown JB, Smith DH (2004). Longitudinal follow-up and outcomes among a population with chronic kidney disease in a large managed care organization. Archives of internal medicine, 164:659-663. - PubMed
-
- Mizushima N, Levine B (2020). Autophagy in Human Diseases. N Engl J Med, 383:1564-1576. - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Research Materials
