Analysis of Whole-Transcriptome RNA-Seq Data Reveals the Involvement of Alternative Splicing in the Drought Response of Glycyrrhiza uralensis
- PMID: 35656323
- PMCID: PMC9152209
- DOI: 10.3389/fgene.2022.885651
Analysis of Whole-Transcriptome RNA-Seq Data Reveals the Involvement of Alternative Splicing in the Drought Response of Glycyrrhiza uralensis
Abstract
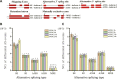
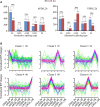
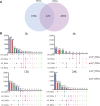
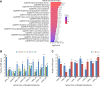
Alternative splicing (AS) is a post-transcriptional regulatory mechanism that increases protein diversity. There is growing evidence that AS plays an important role in regulating plant stress responses. However, the mechanism by which AS coordinates with transcriptional regulation to regulate the drought response in Glycyrrhiza uralensis remains unclear. In this study, we performed a genome-wide analysis of AS events in G. uralensis at different time points under drought stress using a high-throughput RNA sequencing approach. We detected 2,479 and 2,764 AS events in the aerial parts (AP) and underground parts (UP), respectively, of drought-stressed G. uralensis. Of these, last exon AS and exon skipping were the main types of AS. Overall, 2,653 genes undergoing significant AS regulation were identified from the AP and UP of G. uralensis exposed to drought for 2, 6, 12, and 24 h. Gene Ontology analyses indicated that AS plays an important role in the regulation of nitrogen and protein metabolism in the drought response of G. uralensis. Notably, the spliceosomal pathway and basal transcription factor pathway were significantly enriched with differentially spliced genes under drought stress. Genes related to splicing regulators in the AP and UP of G. uralensis responded to drought stress and underwent AS under drought conditions. In summary, our data suggest that drought-responsive AS directly and indirectly regulates the drought response of G. uralensis. Further in-depth studies on the functions and mechanisms of AS during abiotic stresses will provide new strategies for improving plant stress resistance.
Keywords: Glycyrrhiza uralensis; alternative splicing; drought regulators; drought response; splicing regulatory factor; transcriptome analysis.
Copyright © 2022 Li, Xu, Huang, Bi, Yang, Shen and Liu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



Similar articles
-
Alternative splicing responses to salt stress in Glycyrrhiza uralensis revealed by global profiling of transcriptome RNA-seq datasets.Front Genet. 2024 Jul 9;15:1397502. doi: 10.3389/fgene.2024.1397502. eCollection 2024. Front Genet. 2024. PMID: 39045328 Free PMC article.
-
Analysis of Whole Transcriptome RNA-seq Data Reveals Many Alternative Splicing Events in Soybean Roots under Drought Stress Conditions.Genes (Basel). 2020 Dec 19;11(12):1520. doi: 10.3390/genes11121520. Genes (Basel). 2020. PMID: 33352659 Free PMC article.
-
Combined Analysis of Pharmaceutical Active Ingredients and Transcriptomes of Glycyrrhiza uralensis Under PEG6000-Induced Drought Stress Revealed Glycyrrhizic Acid and Flavonoids Accumulation via JA-Mediated Signaling.Front Plant Sci. 2022 Jun 13;13:920172. doi: 10.3389/fpls.2022.920172. eCollection 2022. Front Plant Sci. 2022. PMID: 35769299 Free PMC article.
-
Decoding co-/post-transcriptional complexities of plant transcriptomes and epitranscriptome using next-generation sequencing technologies.Biochem Soc Trans. 2020 Dec 18;48(6):2399-2414. doi: 10.1042/BST20190492. Biochem Soc Trans. 2020. PMID: 33196096 Review.
-
Alternative splicing: enhancing ability to cope with stress via transcriptome plasticity.Plant Sci. 2012 Apr;185-186:40-9. doi: 10.1016/j.plantsci.2011.09.006. Epub 2011 Sep 28. Plant Sci. 2012. PMID: 22325865 Review.
Cited by
-
Alternative splicing responses to salt stress in Glycyrrhiza uralensis revealed by global profiling of transcriptome RNA-seq datasets.Front Genet. 2024 Jul 9;15:1397502. doi: 10.3389/fgene.2024.1397502. eCollection 2024. Front Genet. 2024. PMID: 39045328 Free PMC article.
-
Comprehensive physiological, transcriptomic, and metabolomic analyses reveal the synergistic mechanism of Bacillus pumilus G5 combined with silicon alleviate oxidative stress in drought-stressed Glycyrrhiza uralensis Fisch.Front Plant Sci. 2022 Dec 8;13:1033915. doi: 10.3389/fpls.2022.1033915. eCollection 2022. Front Plant Sci. 2022. PMID: 36570944 Free PMC article.
-
Differential alternative splicing genes and isoform co-expression networks of Brassica napus under multiple abiotic stresses.Front Plant Sci. 2022 Oct 13;13:1009998. doi: 10.3389/fpls.2022.1009998. eCollection 2022. Front Plant Sci. 2022. PMID: 36311064 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Research Materials

