Panel-based next-generation sequencing identifies novel mutations in Bulgarian patients with inherited retinal dystrophies
- PMID: 35656873
- PMCID: PMC9356554
- DOI: 10.1002/mgg3.1997
Panel-based next-generation sequencing identifies novel mutations in Bulgarian patients with inherited retinal dystrophies
Abstract
Background: Next-generation sequencing (NGS)-based method is being used broadly for genetic testing especially for clinically and genetically heterogeneous disorders, such as inherited retinal degenerations (IRDs) but still not routinely used for molecular diagnostics in Bulgaria. Consequently, the purpose of this study was to evaluate the effectiveness of a molecular diagnostic approach, based on targeted NGS for the identification of the disease-causing mutations in 16 Bulgarian patients with different IRDs.
Methods: We applied a customized NGS panel, including 125 genes associated with retinal and other eye diseases to the patients with hereditary retinopathies.
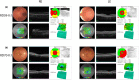
Results: Systematic filtering approach coupled with copy number variation analysis and segregation study lead to the identification of 16 pathogenic and likely pathogenic variants in 12/16 (75%) of IRD patients, 2 of which novel (12.5%): ABCA4-c.668delA (p.K223Rfs18) and RР1-c.2015dupA (p.K673Efs*25). Mutations in the ABCA4, PRPH2, USH2A, BEST1, RР1, CDHR1, and RHO genes were detected reaching a diagnostic yield between 42.9% for Retinitis pigmentosa cases and 100% for macular degeneration, Usher syndrome, and cone-rod dystrophy patients.
Conclusion: Our results confirm the usefulness of targeted NGS approach based on frequently mutated genes as a comprehensive and successful genetic diagnostic tool for IRDs with significant impact on patients counseling.
Keywords: inherited retinal degeneration; molecular diagnostics; novel mutations; targeted next generation sequencing.
© 2022 The Authors. Molecular Genetics & Genomic Medicine published by Wiley Periodicals LLC.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Albert, S. , Garanto, A. , Sangermano, R. , Khan, M. , Bax, N. M. , Hoyng, C. B. , & Cremers, F. P. M. (2018). Identification and rescue of splice defects caused by two neighboring deep‐intronic ABCA4 mutations underlying Stargardt disease. American Journal of Human Genetics, 102(4), 517–527. 10.1016/j.ajhg.2018.02.008 - DOI - PMC - PubMed
-
- Arno, G. , Hull, S. , Carss, K. , Dev Borman, A. , Chakarova, C. , Bujakowska, K. , & Webster, A. (2016). Reevaluation of the retinal dystrophy due to recessive alleles of RGR with the discovery of a cis‐acting mutation in CDHR1. Investigative Ophthalmology & Visual Science, 57, 4806–4813. 10.1167/iovs.16-19687 - DOI - PubMed
-
- Bennett, J. , Wellman, J. , Marshall, K. A. , McCague, S. , Ashtari, M. , DiStefano‐Pappas, J. , & Maguire, A. M. (2016). Safety and durability of effect of contralateral‐eye administration of AAV2 gene therapy in patients with childhood‐onset blindness caused by RPE65 mutations: A follow‐on phase 1 trial. Lancet, 388(10045), 661–672. 10.1016/s0140-6736(16)30371-3 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

