Noncanonical prokaryotic X family DNA polymerases lack polymerase activity and act as exonucleases
- PMID: 35657103
- PMCID: PMC9226535
- DOI: 10.1093/nar/gkac461
Noncanonical prokaryotic X family DNA polymerases lack polymerase activity and act as exonucleases
Abstract
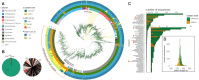
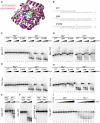
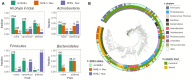
The X family polymerases (PolXs) are specialized DNA polymerases that are found in all domains of life. While the main representatives of eukaryotic PolXs, which have dedicated functions in DNA repair, were studied in much detail, the functions and diversity of prokaryotic PolXs have remained largely unexplored. Here, by combining a comprehensive bioinformatic analysis of prokaryotic PolXs and biochemical experiments involving selected recombinant enzymes, we reveal a previously unrecognized group of PolXs that seem to be lacking DNA polymerase activity. The noncanonical PolXs contain substitutions of the key catalytic residues and deletions in their polymerase and dNTP binding sites in the palm and fingers domains, but contain functional nuclease domains, similar to canonical PolXs. We demonstrate that representative noncanonical PolXs from the Deinococcus genus are indeed inactive as DNA polymerases but are highly efficient as 3'-5' exonucleases. We show that both canonical and noncanonical PolXs are often encoded together with the components of the non-homologous end joining pathway and may therefore participate in double-strand break repair, suggesting an evolutionary conservation of this PolX function. This is a remarkable example of polymerases that have lost their main polymerase activity, but retain accessory functions in DNA processing and repair.
© The Author(s) 2022. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


Similar articles
-
Characterization of DNA polymerase X from Thermus thermophilus HB8 reveals the POLXc and PHP domains are both required for 3'-5' exonuclease activity.Nucleic Acids Res. 2009 Apr;37(6):2037-52. doi: 10.1093/nar/gkp064. Epub 2009 Feb 11. Nucleic Acids Res. 2009. PMID: 19211662 Free PMC article.
-
DNA stabilization at the Bacillus subtilis PolX core--a binding model to coordinate polymerase, AP-endonuclease and 3'-5' exonuclease activities.Nucleic Acids Res. 2012 Oct;40(19):9750-62. doi: 10.1093/nar/gks702. Epub 2012 Jul 25. Nucleic Acids Res. 2012. PMID: 22844091 Free PMC article.
-
A conserved 3'----5' exonuclease active site in prokaryotic and eukaryotic DNA polymerases.Cell. 1989 Oct 6;59(1):219-28. doi: 10.1016/0092-8674(89)90883-0. Cell. 1989. PMID: 2790959
-
Translesion synthesis DNA polymerases and control of genome stability.Front Biosci. 2006 Sep 1;11:2496-517. doi: 10.2741/1985. Front Biosci. 2006. PMID: 16720328 Review.
-
Polymerases in nonhomologous end joining: building a bridge over broken chromosomes.Antioxid Redox Signal. 2011 Jun 15;14(12):2509-19. doi: 10.1089/ars.2010.3429. Epub 2010 Oct 28. Antioxid Redox Signal. 2011. PMID: 20649463 Free PMC article. Review.
Cited by
-
Structural and Evolutionary Analysis of Proteins Endowed with a Nucleotidyltransferase, or Non-canonical Palm, Catalytic Domain.J Mol Evol. 2024 Dec;92(6):799-814. doi: 10.1007/s00239-024-10207-7. Epub 2024 Sep 19. J Mol Evol. 2024. PMID: 39297932 Free PMC article.
-
Fidelity, specialization, and evolution of Paramecium PolX DNA polymerases involved in programmed double-strand break DNA repair.Nucleic Acids Res. 2025 Aug 11;53(15):gkaf786. doi: 10.1093/nar/gkaf786. Nucleic Acids Res. 2025. PMID: 40829804 Free PMC article.
-
Terminal deoxynucleotidyl transferase: Properties and applications.Eng Microbiol. 2024 Nov 28;5(1):100179. doi: 10.1016/j.engmic.2024.100179. eCollection 2025 Mar. Eng Microbiol. 2024. PMID: 40538715 Free PMC article. Review.
References
-
- Raia P., Delarue M., Sauguet L.. An updated structural classification of replicative DNA polymerases. Biochem. Soc. Trans. 2019; 47:239–249. - PubMed
-
- Jain R., Aggarwal A.K., Rechkoblit O.. Eukaryotic DNA polymerases. Curr. Opin. Struct. Biol. 2018; 53:77–87. - PubMed
-
- Baril E.F., Brown O.E., Jenkins M.D., Laszlo J.. Deoxyribonucleic acid polymerase with rat liver ribosomes and smooth membranes. Purification and properties of the enzymes. Biochemistry. 1971; 10:1981–1992. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

