Integrin alpha 6 is upregulated and drives hepatocellular carcinoma progression through integrin α6β4 complex
- PMID: 35657344
- PMCID: PMC9329238
- DOI: 10.1002/ijc.34146
Integrin alpha 6 is upregulated and drives hepatocellular carcinoma progression through integrin α6β4 complex
Abstract
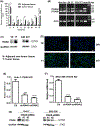
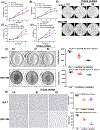
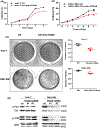
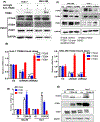
Integrin α6 (ITGA6) forms integrin receptors with either integrin β1 (ITGB1) or integrin β4 (ITGB4). How it functions to regulate hepatocellular carcinoma (HCC) progression is not well-elucidated. We found that ITGA6 RNA and protein expression levels are significantly elevated in human HCC tissues in comparison with paired adjacent nontumor tissues by RNA sequencing, RT-qPCR, Western blotting and immunofluorescence staining. Stable knockdown of ITGA6 with different ITGA6 shRNA expression lentivectors significantly inhibited proliferation, migration and anchorage-independent growth of HCC cell lines in vitro, and xenograft tumor growth in vivo. The inhibition of anchorage-dependent and -independent growth of HCC cell lines was also confirmed with anti-ITGA6 antibody. ITGA6 knockdown was shown to induce cell-cycle arrest at G0/G1 phase. Immunoprecipitation assay revealed apparent interaction of ITGA6 with ITGB4, but not ITGB1. Expression studies showed that ITGA6 positively regulates the expression of ITGB4 with no or negative regulation of ITGB1 expression. Finally, while high levels of ITGA6 and ITGB4 together were associated with significantly worse survival of HCC patients in TCGA data set, the association was not significant for high levels of ITGA6 and ITGB1. In conclusion, ITGA6 is upregulated in HCC tumors and has a malignant promoting role in HCC cells through integrin α6β4 complex. Thus, integrin α6β4 may be a therapeutic target for treating patients with HCC.
Keywords: hepatocellular carcinoma; integrin α6; integrin α6β4; migration; proliferation.
© 2022 UICC.
Conflict of interest statement
CONFLICT OF INTEREST
The authors declare that they have no conflict of interests.
Figures


References
-
- Sia D, Villanueva A, Friedman SL, Llovet JM. Liver Cancer Cell of Origin, Molecular Class, and Effects on Patient Prognosis. Gastroenterology 2017;152: 745–61. - PubMed
-
- Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68: 394–424. - PubMed
-
- El-Serag HB. Hepatocellular carcinoma. N Engl J Med 2011;365: 1118–27. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

