Surface-Induced Dissociation for Protein Complex Characterization
- PMID: 35657596
- PMCID: PMC11892799
- DOI: 10.1007/978-1-0716-2325-1_15
Surface-Induced Dissociation for Protein Complex Characterization
Abstract
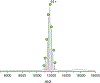
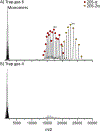
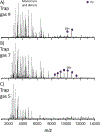
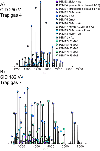
Native mass spectrometry (nMS) enables intact non-covalent complexes to be studied in the gas phase. nMS can provide information on composition, stoichiometry, topology, and, when coupled with surface-induced dissociation (SID), subunit connectivity. Here we describe the characterization of protein complexes by nMS and SID. Substructural information obtained using this method is consistent with the solved complex structure, when a structure exists. This provides confidence that the method can also be used to obtain substructural information for unknowns, providing insight into subunit connectivity and arrangements. High-energy SID can also provide information on proteoforms present. Previously SID has been limited to a few in-house modified instruments and here we focus on SID implemented within an in-house-modified Q Exactive UHMR. However, SID is currently commercially available within the Waters Select Series Cyclic IMS instrument. Projects are underway that involve the NIH-funded native MS resource (nativems.osu.edu), instrument vendors, and third-party vendors, with the hope of bringing the technology to more platforms and labs in the near future. Currently, nMS resource staff can perform SID experiments for interested research groups.
Keywords: High-resolution mass spectrometry; Native mass spectrometry; Protein complex; Protein mass spectrometry; Proteoform identification; Surface-induced dissociation.
© 2022. The Author(s), under exclusive license to Springer Science+Business Media, LLC, part of Springer Nature.
Figures

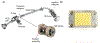
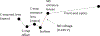

References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

