Emergency SARS-CoV-2 variants of concern: rapidly direct RT-qPCR detection without RNA extraction, clinical comparison, cost-effective, and high-throughput
- PMID: 35657641
- PMCID: PMC9217698
- DOI: 10.18632/aging.204095
Emergency SARS-CoV-2 variants of concern: rapidly direct RT-qPCR detection without RNA extraction, clinical comparison, cost-effective, and high-throughput
Abstract
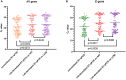
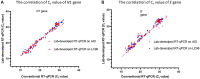
Since the late 2020, the evolution of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variants of concern has been characterized by the emergence of spike protein mutations, and these variants have become dominant worldwide. The gold standard SARS-CoV-2 diagnosis protocol requires two complex processes, namely, RNA extraction and real-time reverse transcriptase polymerase chain reaction (RT-PCR). There is a need for a faster, simpler, and more cost-effective detection strategy that can be utilized worldwide, especially in developing countries. We propose the novel use of direct RT-qPCR, which does not require RNA extraction or a preheating step. For the detection, retrospectively, we used 770 clinical nasopharyngeal swabs, including positive and negative samples. The samples were subjected to RT-qPCR in the N1 and E genes using two different thermocyclers. The limit of detection was 30 copies/reaction for N1 and 60 copies/reaction for E. Analytical sensitivity was assessed for the developed direct RT-qPCR; the sensitivity was 95.69%, negative predictive value was 99.9%, accuracy of 99.35%, and area under the curve was 0.978. This novel direct RT-qPCR diagnosis method without RNA extraction is a reliable and high-throughput alternative method that can significantly save cost, labor, and time during the coronavirus disease 2019 pandemic.
Keywords: COVID-19; SARS-CoV-2 VOC; cost-effective diagnosis; direct RT-qPCR; high-throughput.
Conflict of interest statement
Figures
References
-
- Galloway SE, Paul P, MacCannell DR, Johansson MA, Brooks JT, MacNeil A, Slayton RB, Tong S, Silk BJ, Armstrong GL, Biggerstaff M, Dugan VG. Emergence of SARS-CoV-2 B.1.1.7 Lineage - United States, December 29, 2020-January 12, 2021. MMWR Morb Mortal Wkly Rep. 2021; 70:95–9. 10.15585/mmwr.mm7003e2 - DOI - PMC - PubMed
-
- Chung HY, Jian MJ, Chang CK, Lin JC, Yeh KM, Yang YS, Chen CW, Hsieh SS, Tang SH, Perng CL, Chang FY, Hung KS, Chen ES, et al. Multicenter study evaluating one multiplex RT-PCR assay to detect SARS-CoV-2, influenza A/B, and respiratory syncytia virus using the LabTurbo AIO open platform: epidemiological features, automated sample-to-result, and high-throughput testing. Aging (Albany NY). 2021; 13:24931–42. 10.18632/aging.203761 - DOI - PMC - PubMed
-
- Davies NG, Abbott S, Barnard RC, Jarvis CI, Kucharski AJ, Munday JD, Pearson CAB, Russell TW, Tully DC, Washburne AD, Wenseleers T, Gimma A, Waites W, et al. , and CMMID COVID-19 Working Group, and COVID-19 Genomics UK (COG-UK) Consortium. Estimated transmissibility and impact of SARS-CoV-2 lineage B.1.1.7 in England. Science. 2021; 372:eabg3055. 10.1126/science.abg3055 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

