Potential of antiviral peptide-based SARS-CoV-2 inactivators to combat COVID-19
- PMID: 35657783
- PMCID: PMC9165783
- DOI: 10.1371/journal.pone.0268919
Potential of antiviral peptide-based SARS-CoV-2 inactivators to combat COVID-19
Retraction in
-
Retraction: Potential of antiviral peptide-based SARS-CoV-2 inactivators to combat COVID-19.PLoS One. 2022 Aug 31;17(8):e0273489. doi: 10.1371/journal.pone.0273489. eCollection 2022. PLoS One. 2022. PMID: 36044431 Free PMC article. No abstract available.
Abstract
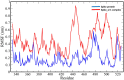
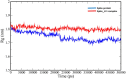
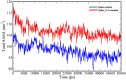
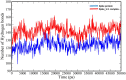
The appearance of new variants of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and the lack of effective antiviral therapeutics for coronavirus disease 2019 (COVID-19), a highly infectious disease caused by the virus, demands the search for alternative therapies. Most antiviral drugs known are passive defenders which must enter the cell to execute their function and suffer from concerns such as permeability and effectiveness, therefore in this current study, we aim to identify peptide inactivators that can act without entering the cells. SARS-CoV-2 spike protein is an essential protein that plays a major role in binding to the host receptor angiotensin-converting enzyme 2 and mediates the viral cell membrane fusion process. SARS vaccines and treatments have also been developed with the spike protein as a target. The virtual screening experiment revealed antiviral peptides which were found to be non-allergen, non-toxic and possess good water solubility. U-1, GST-removed-HR2 and HR2-18 exhibit binding energies of -47.8 kcal/mol, -43.01 kcal/mol, and -40.46 kcal/mol, respectively. The complexes between these peptides and spike protein were stabilized through hydrogen bonds as well as hydrophobic interactions. The stability of the top-ranked peptide with the drug-receptor is evidenced by 50-ns molecular dynamics (MD) simulations. The binding of U-1 induces conformational changes in the spike protein with alterations in its geometric properties such as increased flexibility, decreased compactness, the increased surface area exposed to solvent molecules, and an increase in the number of total hydrogen bonds leading to its probable inactivation. Thus, the identified antiviral peptides can be used as anti-SARS-CoV-2 candidates, inactivating the virus's spike proteins and preventing it from infecting host cells.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

