Temperature Controls eDNA Persistence across Physicochemical Conditions in Seawater
- PMID: 35658125
- PMCID: PMC9231374
- DOI: 10.1021/acs.est.2c01672
Temperature Controls eDNA Persistence across Physicochemical Conditions in Seawater
Abstract
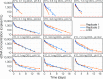
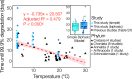
Environmental DNA (eDNA) quantification and sequencing are emerging techniques for assessing biodiversity in marine ecosystems. Environmental DNA can be transported by ocean currents and may remain at detectable concentrations far from its source depending on how long it persist. Thus, predicting the persistence time of eDNA is crucial to defining the spatial context of the information derived from it. To investigate the physicochemical controls of eDNA persistence, we performed degradation experiments at temperature, pH, and oxygen conditions relevant to the open ocean and the deep sea. The eDNA degradation process was best explained by a model with two phases with different decay rate constants. During the initial phase, eDNA degraded rapidly, and the rate was independent of physicochemical factors. During the second phase, eDNA degraded slowly, and the rate was strongly controlled by temperature, weakly controlled by pH, and not controlled by dissolved oxygen concentration. We demonstrate that marine eDNA can persist at quantifiable concentrations for over 2 weeks at low temperatures (≤10 °C) but for a week or less at ≥20 °C. The relationship between temperature and eDNA persistence is independent of the source species. We propose a general temperature-dependent model to predict the maximum persistence time of eDNA detectable through single-species eDNA quantification methods.
Keywords: coral; deep sea; environmental DNA; marine; persistence.
Conflict of interest statement
The authors declare no competing financial interest.
Figures


References
-
- Thomsen P. F.; Willerslev E. Environmental DNA - An Emerging Tool in Conservation for Monitoring Past and Present Biodiversity. Biol. Conserv. 2015, 183, 4–18. 10.1016/j.biocon.2014.11.019. - DOI
-
- Breed M. F.; Harrison P. A.; Blyth C.; Byrne M.; Gaget V.; Gellie N. J. C.; Groom S. V. C.; Hodgson R.; Mills J. G.; Prowse T. A. A.; Steane D. A.; Mohr J. J. The Potential of Genomics for Restoring Ecosystems and Biodiversity. Nat. Rev. Genet. 2019, 20, 615–628. 10.1038/s41576-019-0152-0. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

