A Virus Is a Community: Diversity within Negative-Sense RNA Virus Populations
- PMID: 35658541
- PMCID: PMC9491172
- DOI: 10.1128/mmbr.00086-21
A Virus Is a Community: Diversity within Negative-Sense RNA Virus Populations
Abstract
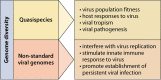
Negative-sense RNA virus populations are composed of diverse viral components that interact to form a community and shape the outcome of virus infections. At the genomic level, RNA virus populations consist not only of a homogeneous population of standard viral genomes but also of an extremely large number of genome variants, termed viral quasispecies, and nonstandard viral genomes, which include copy-back viral genomes, deletion viral genomes, mini viral RNAs, and hypermutated RNAs. At the particle level, RNA virus populations are composed of pleomorphic particles, particles missing or having additional genomes, and single particles or particle aggregates. As we continue discovering more about the components of negative-sense RNA virus populations and their crucial functions during virus infection, it will become more important to study RNA virus populations as a whole rather than their individual parts. In this review, we will discuss what is known about the components of negative-sense RNA virus communities, speculate how the components of the virus community interact, and summarize what vaccines and antiviral therapies are being currently developed to target or harness these components.
Keywords: antiviral immunity; copy-back viral genomes; copy-backs; deletion viral genomes; filamentous particles; multiple genome packaging; negative-sense RNA virus; nonstandard viral genomes; semi-infectious particles; viral quasispecies; virion aggregation; virus; virus community.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

