Simulating the restoration of normal gene expression from different thyroid cancer stages using deep learning
- PMID: 35659616
- PMCID: PMC9166476
- DOI: 10.1186/s12885-022-09704-z
Simulating the restoration of normal gene expression from different thyroid cancer stages using deep learning
Abstract
Background: Thyroid cancer (THCA) is the most common endocrine malignancy and incidence is increasing. There is an urgent need to better understand the molecular differences between THCA tumors at different pathologic stages so appropriate diagnostic, prognostic, and treatment strategies can be applied. Transcriptome State Perturbation Generator (TSPG) is a tool created to identify the changes in gene expression necessary to transform the transcriptional state of a source sample to mimic that of a target.
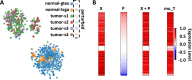
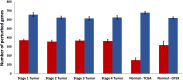
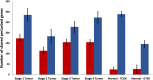
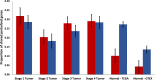
Methods: We used TSPG to perturb the bulk RNA expression data from various THCA tumor samples at progressive stages towards the transcriptional pattern of normal thyroid tissue. The perturbations produced were analyzed to determine if there are consistently up- or down-regulated genes or functions in certain stages of tumors.
Results: Some genes of particular interest were investigated further in previous research. SLC6A15 was found to be down-regulated in all stage 1-3 samples. This gene has previously been identified as a tumor suppressor. The up-regulation of PLA2G12B in all samples was notable because the protein encoded by this gene belongs to the PLA2 superfamily, which is involved in metabolism, a major function of the thyroid gland. REN was up-regulated in all stage 3 and 4 samples. The enzyme renin encoded by this gene, has a role in the renin-angiotensin system; this system regulates angiogenesis and may have a role in cancer development and progression. This is supported by the consistent up-regulation of REN only in later stage tumor samples. Functional enrichment analysis showed that olfactory receptor activities and similar terms were enriched for the up-regulated genes which supports previous research concluding that abundance and stimulation of olfactory receptors is linked to cancer.
Conclusions: TSPG can be a useful tool in exploring large gene expression datasets and extracting the meaningful differences between distinct classes of data. We identified genes that were characteristically perturbed in certain sample types, including only late-stage THCA tumors. Additionally, we provided evidence for potential transcriptional signatures of each stage of thyroid cancer. These are potentially relevant targets for future investigation into THCA tumorigenesis.
Keywords: Deep learning; TSPG; Thyroid cancer; Transcriptome.
© 2022. The Author(s).
Conflict of interest statement
Not applicable.
Figures
Similar articles
-
Identification of hub genes in thyroid carcinoma to predict prognosis by integrated bioinformatics analysis.Bioengineered. 2021 Dec;12(1):2928-2940. doi: 10.1080/21655979.2021.1940615. Bioengineered. 2021. PMID: 34167437 Free PMC article.
-
NRXN2 Possesses a Tumor Suppressor Potential via Inhibiting the Growth of Thyroid Cancer Cells.Comput Math Methods Med. 2021 Nov 3;2021:7993622. doi: 10.1155/2021/7993622. eCollection 2021. Comput Math Methods Med. 2021. PMID: 34777568 Free PMC article.
-
E2F, HSF2, and miR-26 in thyroid carcinoma: bioinformatic analysis of RNA-sequencing data.Genet Mol Res. 2016 Mar 11;15(1):15017576. doi: 10.4238/gmr.15017576. Genet Mol Res. 2016. PMID: 26985959
-
SLC6A15 acts as a tumor suppressor to inhibit migration and invasion in human papillary thyroid cancer.J Cell Biochem. 2021 Aug;122(8):814-826. doi: 10.1002/jcb.29914. Epub 2021 Mar 10. J Cell Biochem. 2021. PMID: 33690923
-
CircHACE1 functions as a competitive endogenous RNA to curb differentiated thyroid cancer progression by upregulating Tfcp2L1 through adsorbing miR-346.Endocr J. 2021 Aug 28;68(8):1011-1025. doi: 10.1507/endocrj.EJ20-0806. Epub 2021 Jun 4. Endocr J. 2021. PMID: 34092745
Cited by
-
GEMDiff: a diffusion workflow bridges between normal and tumor gene expression states: a breast cancer case study.Brief Bioinform. 2025 Mar 4;26(2):bbaf093. doi: 10.1093/bib/bbaf093. Brief Bioinform. 2025. PMID: 40067113 Free PMC article.
References
-
- Wiltshire JJ, Drake TM, Uttley L, Balasubramanian SP. Systematic Review of Trends in the Incidence Rates of Thyroid Cancer. Thyroid (New York, N.Y.). 2016;26(11):1541–52. - PubMed
-
- Howlader N, Noone AM, Krapcho M, Miller D, Brest A, Yu M, Ruhl J, Tatalovich Z, Mariotto A, Lewis DR, Chen HS. SEER Cancer Statistics Review. 1975–2018;2021 ASI 4474–35. 2021.
-
- Cabanillas ME. Dr, McFadden DG, MD, Durante C. MD Thyroid cancer The Lancet (British edition) 2016;388(10061):2783–2795. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

