Structure of Reelin repeat 8 and the adjacent C-terminal region
- PMID: 35659645
- PMCID: PMC9300658
- DOI: 10.1016/j.bpj.2022.06.002
Structure of Reelin repeat 8 and the adjacent C-terminal region
Abstract
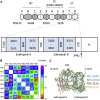
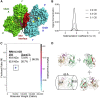
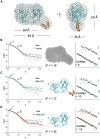
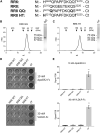
Neuronal development and function are dependent in part on the several roles of the secreted glycoprotein Reelin. Endogenous proteases process this 400 kDa, modular protein, yielding N-terminal, central, and C-terminal fragments that each have distinct roles in Reelin's function and regulation. The C-terminal fragment comprises Reelin repeat (RR) domains seven and eight, as well as a basic stretch of 32 amino acid residues termed the C-terminal region (CTR), influences Reelin signaling intensity, and has been reported to bind to Neuropilin-1, which serves as a co-receptor in the canonical Reelin signaling pathway. Here, we present a crystal structure of RR8 at 3.0 Å resolution. Analytical ultracentrifugation and small-angle x-ray scattering confirmed that RR8 is monomeric and enabled us to identify the CTR as a flexible, yet compact subdomain. We conducted structurally informed protein engineering to design a chimeric RR8 construct guided by the structural similarities with RR6. Experimental results support a mode of Reelin-receptor interaction reliant on the multiple interfaces coordinating the binding event. Structurally, RR8 resembles other individual RRs, but its structure does show discrete differences that may account for Reelin receptor specificity toward RR6.
Copyright © 2022 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures


References
-
- Iafrati J., Orejarena M.J., et al. Chavis P. Reelin, an extracellular matrix protein linked to early onset psychiatric diseases, drives postnatal development of the prefrontal cortex via GluN2B-NMDARs and the mTOR pathway. Mol. Psychiatr. 2014;19:417–426. doi: 10.1038/mp.2013.66. - DOI - PMC - PubMed
-
- Lambert de Rouvroit C., Goffinet A.M. The reeler mouse as a model of brain development. Adv. Anat. Embryol. Cell Biol. 1998;150:1–106. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

