CRISPR detection of circulating cell-free Mycobacterium tuberculosis DNA in adults and children, including children with HIV: a molecular diagnostics study
- PMID: 35659882
- PMCID: PMC9300929
- DOI: 10.1016/S2666-5247(22)00087-8
CRISPR detection of circulating cell-free Mycobacterium tuberculosis DNA in adults and children, including children with HIV: a molecular diagnostics study
Abstract
Background: Tuberculosis remains a leading cause of global mortality, especially for adults and children living with HIV (CLHIV) underdiagnosed by sputum-based assays. Non-sputum-based assays are needed to improve tuberculosis diagnosis and tuberculosis treatment monitoring. Our aim in this study was to determine whether ultrasensitive detection of Mycobacterium tuberculosis cell-free DNA (Mtb-cfDNA) in blood can diagnose tuberculosis and evaluate tuberculosis treatment responses.
Methods: In this molecular diagnostics study we analysed archived serum from two patient populations evaluated for tuberculosis in Eswatini and Kenya to detect Mtb-cfDNA, analysing serum from all individuals who had both sufficient serum volumes and clear diagnostic results. An optimised CRISPR-mediated tuberculosis (CRISPR-TB) assay was used to detect Mtb-cfDNA in serum at enrolment from adults and children with presumptive tuberculosis and their asymptomatic household contacts, and at enrolment and during tuberculosis treatment from a cohort of symptomatic CLHIV at high risk for tuberculosis, who provided longitudinal serum at enrolment and during tuberculosis treatment.
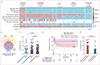
Findings: CRISPR-TB identified microbiologically and clinically confirmed tuberculosis cases in the predominantly HIV-negative Eswatini adult cohort with 96% sensitivity (27 [96%] of 28, 95% CI 80-100) and 94% specificity (16 [94%] of 17, 71-100), and with 83% sensitivity (5 [83%] of 6, 36-100) and 95% specificity (21 [95%] of 22, 77-100) in the paediatric cohort, including all six cases of extrapulmonary tuberculosis. In the Kenyan CLHIV cohort, CRISPR-TB detected all (13 [100%] of 13, 75-100) confirmed tuberculosis cases and 85% (39 [85%] of 46, 71-94) of unconfirmed tuberculosis cases diagnosed by non-microbiological clinical findings. CLHIV who were CRISPR-TB positive at enrolment had a 2·4-times higher risk of mortality by 6 months after enrolment. Mtb-cfDNA signal decreased after tuberculosis treatment initiation, with near or complete Mtb-cfDNA clearance by 6 months after tuberculosis treatment initiation.
Interpretation: CRISPR-mediated detection of circulating Mtb-cfDNA shows promise to increase the identification of paediatric tuberculosis and HIV-associated tuberculosis, and potential for early diagnosis and rapid monitoring of tuberculosis treatment responses.
Funding: US Department of Defense, National Institute of Child Health and Human Development, National Institute of Allergy and Infectious Diseases, University of Washington Center for AIDS Research, and the Weatherhead Presidential Endowment fund.
Copyright © 2022 The Author(s). Published by Elsevier Ltd. This is an Open Access article under the CC BY-NC-ND 4.0 license. Published by Elsevier Ltd.. All rights reserved.
Conflict of interest statement
Declaration of interests SML, LMC, GJ-S, and TYH report grants from the NIH. TYH reports grands from DOD and Weatherhead Endowment Fund of Tulane University, during the conduct of the study. JS reports a grant from CFA. SML reports grants from CDC and Merck. TYH, BN, and ZH are inventors on a provisional patent application related to this work filed by Tulane University (62/971210; PCT/US21/16931). All other authors declare no competing interests.
Figures

References
-
- WHO. Global tuberculosis report 2020. Geneva: World Health Organization, 2020.
-
- MacLean E, Broger T, Yerlikaya S, Fernandez-Carballo BL, Pai M, Denkinger CM. A systematic review of biomarkers to detect active tuberculosis. Nat Microbiol 2019; 4: 748–58. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 AI162152/AI/NIAID NIH HHS/United States
- U01 CA252965/CA/NCI NIH HHS/United States
- K23 AI143479/AI/NIAID NIH HHS/United States
- R01 AI122932/AI/NIAID NIH HHS/United States
- R01 AI175618/AI/NIAID NIH HHS/United States
- R21 AI143341/AI/NIAID NIH HHS/United States
- K23 AI120793/AI/NIAID NIH HHS/United States
- R01 HD090927/HD/NICHD NIH HHS/United States
- P30 AI027757/AI/NIAID NIH HHS/United States
- R25 TW011212/TW/FIC NIH HHS/United States
- K23 AI141681/AI/NIAID NIH HHS/United States
- R01 HD103511/HD/NICHD NIH HHS/United States
- R01 AI144168/AI/NIAID NIH HHS/United States
- K01 TW011482/TW/FIC NIH HHS/United States
- R01 HD023412/HD/NICHD NIH HHS/United States
- R01 AI113725/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical

