Cyclin-dependent Kinase 9 as a Potential Target for Anti-TNF-resistant Inflammatory Bowel Disease
- PMID: 35660024
- PMCID: PMC9356186
- DOI: 10.1016/j.jcmgh.2022.05.011
Cyclin-dependent Kinase 9 as a Potential Target for Anti-TNF-resistant Inflammatory Bowel Disease
Abstract
Background & aims: Resistance to single cytokine blockade, namely anti-tumor necrosis factor (TNF) therapy, is a growing concern for patients with inflammatory bowel disease (IBD). The transcription factor T-bet is a critical regulator of intestinal homeostasis, is genetically linked to mucosal inflammation and controls the expression of multiples genes such as the pro-inflammatory cytokines interferon (IFN)-γ and TNF. Inhibiting T-bet may therefore offer a more attractive prospect for treating IBD but remains challenging to target therapeutically. In this study, we evaluate the effect of targeting the transactivation function of T-bet using inhibitors of P-TEFb (CDK9-cyclin T), a transcriptional elongation factor downstream of T-bet.
Methods: Using an adaptive immune-mediated colitis model, human colonic lymphocytes from patients with IBD and multiple large clinical datasets, we investigate the effect of cyclin-dependent kinase 9 (CDK9) inhibitors on cytokine production and gene expression in colonic CD4+ T cells and link these genetic modules to clinical response in patients with IBD.
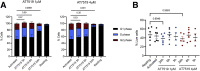
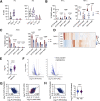
Results: Systemic CDK9 inhibition led to histological improvement of immune-mediated colitis and was associated with targeted suppression of colonic CD4+ T cell-derived IFN-γ and IL-17A. In colonic lymphocytes from patients with IBD, CDK9 inhibition potently repressed genes responsible for pro-inflammatory signalling, and in particular genes regulated by T-bet. Remarkably, CDK9 inhibition targeted genes that were highly expressed in anti-TNF resistant IBD and that predicted non-response to anti-TNF therapy.
Conclusion: Collectively, our findings reveal CDK9 as a potential target for anti-TNF-resistant IBD, which has the potential for rapid translation to the clinic.
Keywords: CDK9; Crohn’s Disease; IBD; Inflammation; Ulcerative Colitis.
Copyright © 2022 The Authors. Published by Elsevier Inc. All rights reserved.
Figures






References
-
- Baumgart D.C., Sandborn W.J. Crohn’s disease. Lancet. 2012;380:1590–1605. - PubMed
-
- Ordás I., Eckmann L., Talamini M., Baumgart D.C., Sandborn W.J. Ulcerative colitis. Lancet. 2012;380:1606–1619. - PubMed
-
- Neurath M.F. Current and emerging therapeutic targets for IBD. Nat Rev Gastroenterol Hepatol. 2017;14:269–278. - PubMed
-
- D’Haens G.R., van Deventer S. 25 years of anti-TNF treatment for inflammatory bowel disease: lessons from the past and a look to the future. Gut. 2021;70:1396–1405. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

