RNF219 regulates CCR4-NOT function in mRNA translation and deadenylation
- PMID: 35660762
- PMCID: PMC9166816
- DOI: 10.1038/s41598-022-13309-8
RNF219 regulates CCR4-NOT function in mRNA translation and deadenylation
Abstract
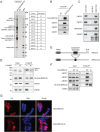
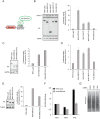
Post-transcriptional regulatory mechanisms play a role in many biological contexts through the control of mRNA degradation, translation and localization. Here, we show that the RING finger protein RNF219 co-purifies with the CCR4-NOT complex, the major mRNA deadenylase in eukaryotes, which mediates translational repression in both a deadenylase activity-dependent and -independent manner. Strikingly, RNF219 both inhibits the deadenylase activity of CCR4-NOT and enhances its capacity to repress translation of a target mRNA. We propose that the interaction of RNF219 with the CCR4-NOT complex directs the translational repressive activity of CCR4-NOT to a deadenylation-independent mechanism.
© 2022. The Author(s).
Conflict of interest statement
A.-R. C. is a member of the scientific advisory board for Flagship Labs 69, Inc.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

