Orb-dependent polyadenylation contributes to PLP expression and centrosome scaffold assembly
- PMID: 35661190
- PMCID: PMC9340551
- DOI: 10.1242/dev.200426
Orb-dependent polyadenylation contributes to PLP expression and centrosome scaffold assembly
Abstract
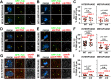
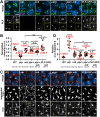
As the microtubule-organizing centers of most cells, centrosomes engineer the bipolar mitotic spindle required for error-free mitosis. Drosophila Pericentrin-like protein (PLP) directs formation of a pericentriolar material (PCM) scaffold required for PCM organization and microtubule-organizing center function. Here, we investigate the post-transcriptional regulation of Plp mRNA. We identify conserved binding sites for cytoplasmic polyadenylation element binding (CPEB) proteins within the Plp 3'-untranslated region and examine the role of the CPEB ortholog Oo18 RNA-binding protein (Orb) in Plp mRNA regulation. Our data show that Orb interacts biochemically with Plp mRNA to promote polyadenylation and PLP protein expression. Loss of orb, but not orb2, diminishes PLP levels in embryonic extracts. Consequently, PLP localization to centrosomes and its function in PCM scaffolding are compromised in orb mutant embryos, resulting in genomic instability and embryonic lethality. Moreover, we find that PLP overexpression restores centrosome scaffolding and rescues the cell division defects caused by orb depletion. Our data suggest that Orb modulates PLP expression at the level of Plp mRNA polyadenylation and demonstrates that the post-transcriptional regulation of core, conserved centrosomal mRNAs is crucial for centrosome function.
Keywords: CPEB; Centrosome; PCM; Polyadenylation; Post-transcriptional regulation; RNA localization.
© 2022. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interests The authors declare no competing or financial interests.
Figures






References
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

