HIF-1-dependent heme synthesis promotes gemcitabine resistance in human non-small cell lung cancers via enhanced ABCB6 expression
- PMID: 35661930
- PMCID: PMC11072486
- DOI: 10.1007/s00018-022-04360-9
HIF-1-dependent heme synthesis promotes gemcitabine resistance in human non-small cell lung cancers via enhanced ABCB6 expression
Abstract
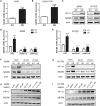
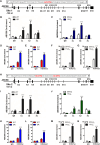
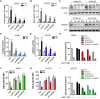
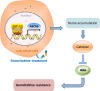
Gemcitabine is commonly used to treat various cancer types, including human non-small cell lung cancer (NSCLC). However, even cases that initially respond rapidly commonly develop acquired resistance, limiting our ability to effectively treat advanced NSCLC. To gain insight for developing a strategy to overcome gemcitabine resistance, the present study investigated the mechanism of gemcitabine resistance in NSCLC according to the involvement of ATP-binding cassette subfamily B member 6 (ABCB6) and heme biosynthesis. First, an analysis of ABCB6 expression in human NSCLCs was found to be associated with poor prognosis and gemcitabine resistance in a hypoxia-inducible factor (HIF)-1-dependent manner. Further experiments showed that activation of HIF-1α/ABCB6 signaling led to intracellular heme metabolic reprogramming and a corresponding increase in heme biosynthesis to enhance the activation and accumulation of catalase. Increased catalase levels diminished the effective levels of reactive oxygen species, thereby promoting gemcitabine-based resistance. In a mouse NSCLC model, inhibition of HIF-1α or ABCB6, in combination with gemcitabine, strongly restrained tumor proliferation, increased tumor cell apoptosis, and prolonged animal survival. These results suggest that, in combination with gemcitabine-based chemotherapy, targeting HIF-1α/ABCB6 signaling could result in enhanced tumor chemosensitivity and, thus, may improve outcomes in NSCLC patients.
Keywords: ABCB6; Gemcitabine resistance; Hypoxia-inducible factor-1; Non-small cell lung cancer; Transcriptional activation.
© 2022. The Author(s), under exclusive licence to Springer Nature Switzerland AG.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



Similar articles
-
Mitochondrial ROS drive resistance to chemotherapy and immune-killing in hypoxic non-small cell lung cancer.J Exp Clin Cancer Res. 2022 Aug 11;41(1):243. doi: 10.1186/s13046-022-02447-6. J Exp Clin Cancer Res. 2022. PMID: 35953814 Free PMC article.
-
Rg3 inhibits gemcitabine-induced lung cancer cell invasiveness through ROS-dependent, NF-κB- and HIF-1α-mediated downregulation of PTX3.J Cell Physiol. 2019 Jul;234(7):10680-10697. doi: 10.1002/jcp.27731. Epub 2019 Jan 9. J Cell Physiol. 2019. PMID: 30628067
-
Propofol suppresses LPS-induced nuclear accumulation of HIF-1α and tumor aggressiveness in non-small cell lung cancer.Oncol Rep. 2017 May;37(5):2611-2619. doi: 10.3892/or.2017.5514. Epub 2017 Mar 17. Oncol Rep. 2017. PMID: 28426124 Free PMC article.
-
Status of hypoxia-inducible factor-1α expression in non-small cell lung cancer.Pharmazie. 2021 Sep 1;76(9):404-411. doi: 10.1691/ph.2021.1524. Pharmazie. 2021. PMID: 34481529 Review.
-
Intracellular trafficking of porphyrins.ACS Chem Biol. 2006 Nov 21;1(10):627-9. doi: 10.1021/cb600442b. ACS Chem Biol. 2006. PMID: 17168567 Free PMC article. Review.
Cited by
-
The role of mitochondrial dysfunction in the cytotoxic synergistic effect of gemcitabine and arsenic on breast cancer.PLoS One. 2025 Jan 7;20(1):e0312424. doi: 10.1371/journal.pone.0312424. eCollection 2025. PLoS One. 2025. PMID: 39774458 Free PMC article.
-
Nanomedicines for Overcoming Cancer Drug Resistance.Pharmaceutics. 2022 Aug 1;14(8):1606. doi: 10.3390/pharmaceutics14081606. Pharmaceutics. 2022. PMID: 36015232 Free PMC article. Review.
-
KIF20A is associated with clinical prognosis and synergistic effect of gemcitabine combined with ferroptosis inducer in lung adenocarcinoma.Front Pharmacol. 2022 Sep 26;13:1007429. doi: 10.3389/fphar.2022.1007429. eCollection 2022. Front Pharmacol. 2022. PMID: 36225575 Free PMC article.
-
A mitochondrial-related gene signature predicts prognosis and immunotherapy response in hepatocellular carcinoma.Sci Rep. 2025 Jul 14;15(1):25435. doi: 10.1038/s41598-025-09464-3. Sci Rep. 2025. PMID: 40659697 Free PMC article.
-
Exploring RNA cargo in extracellular vesicles for pleural mesothelioma detection.BMC Cancer. 2025 Feb 7;25(1):212. doi: 10.1186/s12885-025-13617-y. BMC Cancer. 2025. PMID: 39920655 Free PMC article.
References
MeSH terms
Substances
Grants and funding
- 2017ZX09304023/the National Science and Technology Major Project
- cstc2018jcyjAX0512/the Natural Science Foundation of Chongqing
- cstc2019jcyj-msxmX0791/the Natural Science Foundation of Chongqing
- 2020YJ0046/the Sichuan Science and Technology Program
- 2020YFS0272/the Sichuan Science and Technology Program
LinkOut - more resources
Full Text Sources
Medical

