Development of a dedicated Golden Gate Assembly Platform (RtGGA) for Rhodotorula toruloides
- PMID: 35662893
- PMCID: PMC9157227
- DOI: 10.1016/j.mec.2022.e00200
Development of a dedicated Golden Gate Assembly Platform (RtGGA) for Rhodotorula toruloides
Abstract
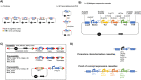
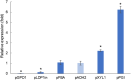
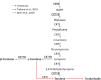
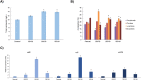
Rhodotorula toruloides is a potential chassis for microbial cell factories as this yeast can metabolise different substrates into a diverse range of natural products, but the lack of efficient synthetic biology tools hinders its applicability. In this study, the modular, versatile and efficient Golden Gate DNA assembly system (RtGGA) was adapted to the first basidiomycete, an oleaginous yeast R. toruloides. R. toruloides CCT 0783 was sequenced, and used for the GGA design. The DNA fragments were assembled with predesigned 4-nt overhangs and a library of standardized parts was created containing promoters, genes, terminators, insertional regions, and resistance genes. The library was combined to create cassettes for the characterization of promoters strength and to overexpress the carotenoid production pathway. A variety of reagents, plasmids, and strategies were used and the RtGGA proved to be robust. The RtGGA was used to build three versions of the carotenoid overexpression cassette by using different promoter combinations. The cassettes were transformed into R. toruloides and the three new strains were characterized. Total carotenoid concentration increased by 41%. The dedicated GGA platform fills a gap in the advanced genome engineering toolkit for R. toruloides, enabling the efficient design of complex metabolic pathways.
Keywords: Golden gate assembly; Metabolic engineering; Non-conventional yeast; Oleaginous yeast; Rhodotorula toruloides; Synthetic biology.
© 2022 The Authors. Published by Elsevier B.V. on behalf of International Metabolic Engineering Society.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures
References
-
- Bankevich A., Nurk S., Antipov D., Gurevich A.A., Dvorkin M., Kulikov A.S., Lesin V.M., Nikolenko S.I., Pham S., Prjibelski A.D., Pyshkin A.V., Sirotkin A.V., Vyahhi N., Tesler G., Alekseyev M.A., Pevzner P.A. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. - DOI - PMC - PubMed
-
- Bonturi N., Crucello A., Viana A.J.C., Miranda E.A. Microbial oil production in sugarcane bagasse hemicellulosic hydrolysate without nutrient supplementation by a Rhodosporidium toruloides adapted strain. Process Biochem. 2017;57:16–25. doi: 10.1016/j.procbio.2017.03.007. - DOI
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

