Reduced serum high-density lipoprotein cholesterol levels and aberrantly expressed cholesterol metabolism genes in colorectal cancer
- PMID: 35663062
- PMCID: PMC9125299
- DOI: 10.12998/wjcc.v10.i14.4446
Reduced serum high-density lipoprotein cholesterol levels and aberrantly expressed cholesterol metabolism genes in colorectal cancer
Abstract
Background: Colorectal cancer (CRC) is a common malignant tumor of the gastrointestinal tract. Lipid metabolism, as an important part of material and energy circulation, is well known to play a crucial role in CRC.
Aim: To explore the relationship between serum lipids and CRC development and identify aberrantly expressed cholesterol metabolism genes in CRC.
Methods: We retrospectively collected 843 patients who had confirmed CRC and received surgical resection from 2013 to 2015 at the Cancer Hospital of the Chinese Academy of Medical Sciences as our research subjects. The levels of serum total cholesterol (TC), triglycerides, low-density lipoprotein cholesterol (LDL-C), high-density lipoprotein cholesterol (HDL-C), LDL-C/HDL-C and clinical features were collected and statistically analyzed by SPSS. Then, we used the data from Oncomine to screen the differentially expressed genes (DEGs) of the cholesterol metabolism pathway in CRC and used Gene Expression Profiling Interactive Analysis to confirm the candidate DEGs. PrognoScan was used to analyze the prognostic value of the DEGs, and Search Tool for the Retrieval of Interacting Genes was used to construct the protein-protein interaction network of DEGs.
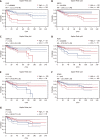
Results: The serum HDL-C level in CRC patients was significantly correlated with tumor size, and patients whose tumor size was more than 5 cm had a lower serum HDL-C level (1.18 ± 0.41 mmol/L vs 1.25 ± 0.35 mmol/L, P < 0.01) than their counterparts. In addition, TC/HDL (4.19 ± 1.33 vs 3.93 ± 1.26, P < 0.01) and LDL-C/HDL-C (2.83 ± 1.10 vs 2.61 ± 0.96, P < 0.01) were higher in patients with larger tumors. The levels of HDL-C (P < 0.05), TC/HDL-C (P < 0.01) and LDL-C/HDL-C (P < 0.05) varied in different stages of CRC patients, and the differences were significant. We screened 14 differentially expressed genes (DEGs) of the cholesterol metabolism pathway in CRC and confirmed that lipoprotein receptor-related protein 8 (LRP8), PCSK9, low-density lipoprotein receptor (LDLR), MBTPS2 and FDXR are upregulated, while ABCA1 and OSBPL1A are downregulated in cancer tissue. Higher expression of LDLR (HR = 3.12, 95%CI: 1.77-5.49, P < 0.001), ABCA1 (HR = 1.66, 95%CI: 1.11-2.48, P = 0.012) and OSBPL1A (HR = 1.38, 95%CI: 1.01-1.89, P = 0.041) all yielded significantly poorer DFS outcomes. Higher expression of FDXR (HR = 0.7, 95%CI: 0.47-1.05, P = 0.002) was correlated with longer DFS. LDLR, ABCA1, OSBPL1A and FDXR were involved in many important cellular function pathways.
Conclusion: Serum HDL-C levels are associated with tumor size and stage in CRC patients. LRP8, PCSK9, LDLR, MBTPS2 and FDXR are upregulated, while ABCA1 and OSBPL1A are downregulated in CRC. Among them, LDLR, ABCA1, OSBPL1A and FDXR were valuable prognostic factors of DFS and were involved in important cellular function pathways.
Keywords: Cholesterol metabolism; Colorectal cancer; High-density lipoprotein cholesterol; Prognosis.
©The Author(s) 2022. Published by Baishideng Publishing Group Inc. All rights reserved.
Conflict of interest statement
Conflict-of-interest statement: The authors declare that there are no conflicts of interest related to this study.
Figures



Similar articles
-
Elevated levels of total cholesterol and high-density lipoprotein cholesterol are associated with better prognosis in patients with lung adenocarcinoma: a retrospective cohort study.Transl Cancer Res. 2025 Apr 30;14(4):2381-2394. doi: 10.21037/tcr-24-1062. Epub 2025 Mar 23. Transl Cancer Res. 2025. PMID: 40386255 Free PMC article.
-
Associations of preoperative serum high-density lipoprotein cholesterol and low-density lipoprotein cholesterol levels with the prognosis of ovarian cancer.Arch Gynecol Obstet. 2022 Mar;305(3):683-691. doi: 10.1007/s00404-021-06215-3. Epub 2021 Aug 28. Arch Gynecol Obstet. 2022. PMID: 34453586
-
Predictive value of a reduction in the level of high-density lipoprotein-cholesterol in patients with non-small-cell lung cancer undergoing radical resection and adjuvant chemotherapy: a retrospective observational study.Lipids Health Dis. 2021 Sep 20;20(1):109. doi: 10.1186/s12944-021-01538-1. Lipids Health Dis. 2021. PMID: 34544437 Free PMC article.
-
Relationship between serum lipid level and colorectal cancer: a systemic review and meta-analysis.BMJ Open. 2022 Jun 22;12(6):e052373. doi: 10.1136/bmjopen-2021-052373. BMJ Open. 2022. PMID: 35732386 Free PMC article.
-
Effects of α-linolenic acid intake on blood lipid profiles:a systematic review and meta-analysis of randomized controlled trials.Crit Rev Food Sci Nutr. 2021;61(17):2894-2910. doi: 10.1080/10408398.2020.1790496. Epub 2020 Jul 9. Crit Rev Food Sci Nutr. 2021. PMID: 32643951
Cited by
-
Mutant APC reshapes Wnt signaling plasma membrane nanodomains by altering cholesterol levels via oncogenic β-catenin.Nat Commun. 2023 Jul 19;14(1):4342. doi: 10.1038/s41467-023-39640-w. Nat Commun. 2023. PMID: 37468468 Free PMC article.
-
Exosomal miR-1470 is a diagnostic biomarker and promotes cell proliferation and metastasis in colorectal cancer.Cancer Med. 2024 Apr;13(7):e7117. doi: 10.1002/cam4.7117. Cancer Med. 2024. PMID: 38545812 Free PMC article.
-
Targeting proprotein convertase subtilisin/kexin type 9 (PCSK9): from bench to bedside.Signal Transduct Target Ther. 2024 Jan 8;9(1):13. doi: 10.1038/s41392-023-01690-3. Signal Transduct Target Ther. 2024. PMID: 38185721 Free PMC article. Review.
-
Dysregulated Cholesterol Metabolism and Its Impact on the Tumour Immune Response in Colorectal Cancer.J Cell Mol Med. 2025 Aug;29(15):e70789. doi: 10.1111/jcmm.70789. J Cell Mol Med. 2025. PMID: 40785042 Free PMC article. Review.
-
Lipid Metabolism-Related Gene Signature Predicts Prognosis and Indicates Immune Microenvironment Infiltration in Advanced Gastric Cancer.Gastroenterol Res Pract. 2024 Feb 26;2024:6639205. doi: 10.1155/2024/6639205. eCollection 2024. Gastroenterol Res Pract. 2024. PMID: 38440405 Free PMC article.
References
-
- Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. - PubMed
-
- Barrington WE, Schenk JM, Etzioni R, Arnold KB, Neuhouser ML, Thompson IM Jr, Lucia MS, Kristal AR. Difference in Association of Obesity With Prostate Cancer Risk Between US African American and Non-Hispanic White Men in the Selenium and Vitamin E Cancer Prevention Trial (SELECT) JAMA Oncol. 2015;1:342–349. - PMC - PubMed
-
- Pastushenko I, Mauri F, Song Y, de Cock F, Meeusen B, Swedlund B, Impens F, Van Haver D, Opitz M, Thery M, Bareche Y, Lapouge G, Vermeersch M, Van Eycke YR, Balsat C, Decaestecker C, Sokolow Y, Hassid S, Perez-Bustillo A, Agreda-Moreno B, Rios-Buceta L, Jaen P, Redondo P, Sieira-Gil R, Millan-Cayetano JF, Sanmatrtin O, D'Haene N, Moers V, Rozzi M, Blondeau J, Lemaire S, Scozzaro S, Janssens V, De Troya M, Dubois C, Pérez-Morga D, Salmon I, Sotiriou C, Helmbacher F, Blanpain C. Fat1 deletion promotes hybrid EMT state, tumour stemness and metastasis. Nature. 2021;589:448–455. - PMC - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

