Epidemiologic and Genomic Analysis of the Severe Acute Respiratory Syndrome Coronavirus 2 Epidemic in the Nebraska Region of the United States, March 2020-2021
- PMID: 35663859
- PMCID: PMC9158493
- DOI: 10.3389/fmicb.2022.878342
Epidemiologic and Genomic Analysis of the Severe Acute Respiratory Syndrome Coronavirus 2 Epidemic in the Nebraska Region of the United States, March 2020-2021
Abstract
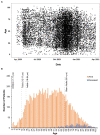
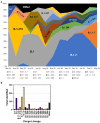
COVID-19 emerged at varying intervals in different regions of the United States in 2020. This report details the epidemiologic and genetic evolution of Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) during the first year of the epidemic in the state of Nebraska using data collected from the Creighton Catholic Health Initiatives (CHI) health system. Statistical modelling identified age, gender, and previous history of diabetes and/or stroke as significant risk factors associated with mortality in COVID-19 patients. In parallel, the viral genomes of over 1,000 samples were sequenced. The overall rate of viral variation in the population was 0.07 mutations/day. Genetically, the first 9 months of the outbreak, which include the initial outbreak, a small surge in August and a major outbreak in November 2020 were primarily characterized by B.1. lineage viruses. In early 2021, the United Kingdom variant (B.1.1.7 or alpha) quickly became the dominant variant. Notably, surveillance of non-consensus variants detected B.1.1.7 defining mutations months earlier in Fall 2020. This work provides insights into the regional variance and evolution of SARS-CoV-2 in the Nebraska region during the first year of the pandemic.
Keywords: COVID-19; SARS-CoV-2; epidemiology; genetic variation; sequencing; viral evolution.
Copyright © 2022 Siedlik, Watson, Raine, Cheng, Goering, Stessman and Belshan.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures








References
-
- Betti M., Bertolotti M., Ferrante D., Roveta A., Pelazza C., Giacchero F., et al. . (2021). Baseline clinical characteristics and prognostic factors in hospitalized COVID-19 patients aged </= 65 years: a retrospective observational study. PLoS One 16:e0248829. doi: 10.1371/journal.pone.0248829, PMID: - DOI - PMC - PubMed
-
- CDC (2021). National Diabetes Statistics Report Website [Online]. Centers for Disease Control and Prevention. Available at: https://www.cdc.gov/diabetes/data/statistics-report/index.html (Accessed December 12, 2021).
-
- Chan J. F.-W., Kok K.-H., Zhu Z., Chu H., To K. K.-W., Yuan S., et al. . (2020). Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg. Microb. Infect. 9, 221–236. doi: 10.1080/22221751.2020.1719902, PMID: - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

