PTBP1 knockdown promotes neural differentiation of glioblastoma cells through UNC5B receptor
- PMID: 35664063
- PMCID: PMC9131277
- DOI: 10.7150/thno.71100
PTBP1 knockdown promotes neural differentiation of glioblastoma cells through UNC5B receptor
Abstract
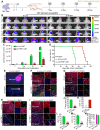
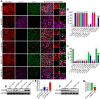
Rationale: Cell reprogramming technology is utilized to prevent cancer progression by transforming cells into terminally differentiated, non-proliferating states. Polypyrimidine tract binding protein 1 (PTBP1) is an RNA binding protein required for the growth of neurons and may directly transform multiple normal human cells into functioning neurons in vitro and in vivo when expressed at low levels. As a result, we identified it as a key to inhibiting cancer cell proliferation by boosting glioblastoma cell neural differentiation. Methods: Immunocytofluorescence (ICF) targeting TUJ1, MAP2, KI67, and EdU were utilized to evaluate glioblastoma cell reprogramming under PTBP1 knockdown or other conditions. PTBP1 and other target genes were detected using Western blotting and qRT-PCR. Activating protein phosphatase 2A (PP2A) and RhoA were detected using specific kits. CCK8 assays were employed to detect cell viability. Bioluminescence, immunohistofluorescence (IHF), and Kaplan-Meier survival analyses were utilized to demonstrate the in vivo reprogramming efficiency of PTBP1 knockdown in U87 murine glioblastoma model. In this study, RNA-seq technology was used to examine the intrinsic pathway. Results: The expression of TUJ1 and MAP2 neural markers, as well as the absence of KI67 and EdU proliferative markers in U251, U87, and KNS89 cells, indicated that glioblastoma cell reprogramming was successful. In vivo, U87 growth generated xenografts was substantially shrank due to PTBP1 knockdown induced neural differentiation, and these tumor-bearing mice had a prolonged survival time. Following RNA-seq, ten potential downstream genes were eliminated. Lentiviral interference and inhibitors blocking tests demonstrated that UNC5B receptor and its downstream signaling were essential in the neural differentiation process mediated by PTBP1 knockdown in glioblastoma cells. Conclusions: Our results indicate that PTBP1 knockdown promotes neural differentiation of glioblastoma cells via UNC5B receptor, consequently suppressing cancer cell proliferation in vitro and in vivo, providing a promising and feasible approach for glioblastoma treatment.
Keywords: PTBP1; cell reprogramming; glioblastoma; neural differentiation; proliferation.
© The author(s).
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures






References
-
- Tan AC, Ashley DM, López GY, Malinzak M, Friedman HS, Khasraw M. Management of glioblastoma: State of the art and future directions. CA Cancer J Clin. 2020;70:299–312. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

